MUCIC - Masaryk University Cell Image Collection
In this web page, we introduce a collection of benchmark data that were generated either synthetically by a computer or created manually. In this sence, the collection serves as a complement to the existing benchmark datasets as it is commonly understood that the validation requires both testing sets: the real and the synthetic one. The real datasets with annotated ground truth offer a realistic variability that is difficult to reach in the synthetic data. Vice versa, the synthetic data can be generated in huge amount and the parameters of the acquisition process can be repeatedly changed with no additional costs. All the generated data are supported by the journal or conference papers.
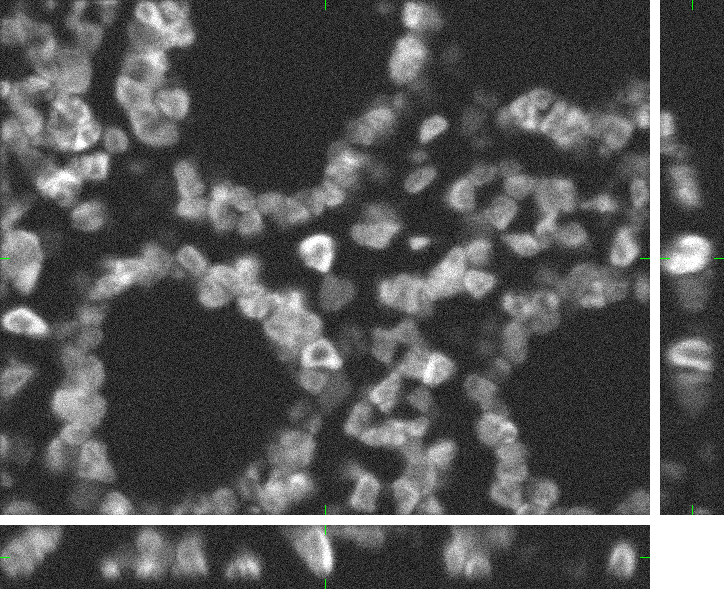
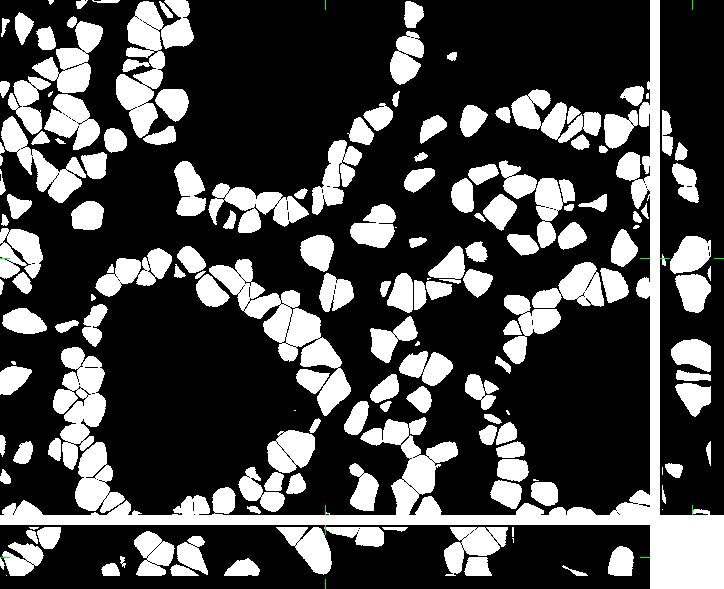
Colon Tissue (fixed cells)
Here, you can find 30 synthetic images of human colon tissue including ground truth (foreground/background) images. The dataset was generated using the virtual microscope imitating the microscope Zeiss S100 (objective Zeiss 63x/1.40 Oil DIC) attached to confocal unit Atto CARV and CCD camera Micromax 1300-YHS. The image data was saved using three different file formats: ICS, HDF5 and 3D-TIFF. Please, feel free to select the format you prefer. All of them contain the same data.
Example images:
 |
 |
|
| 3D image | 3D mask |
The individual image files are aggregated in ZIP archives:
If you use this dataset in your research papers, please refer the following article:
- Svoboda D., Homola O., Stejskal S. Generation of 3D Digital Phantoms of Colon Tissue, In International Conference on Image Analysis and Recognition - ICIAR 2011, Part II, LNCS 6754, Berlin, Heidelberg: Springer-Verlag, pp 31-39, June 2011, ISBN 978-3-642-21595-7
This research was supported by the Ministry of Education of the Czech Republic (Projects No. LC535 and No. 2B06052).
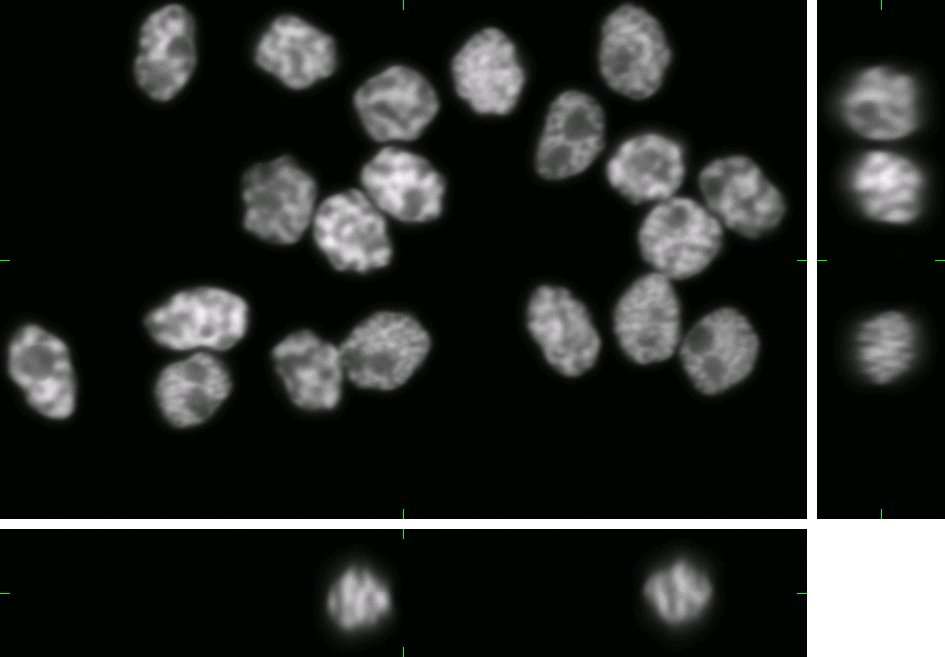
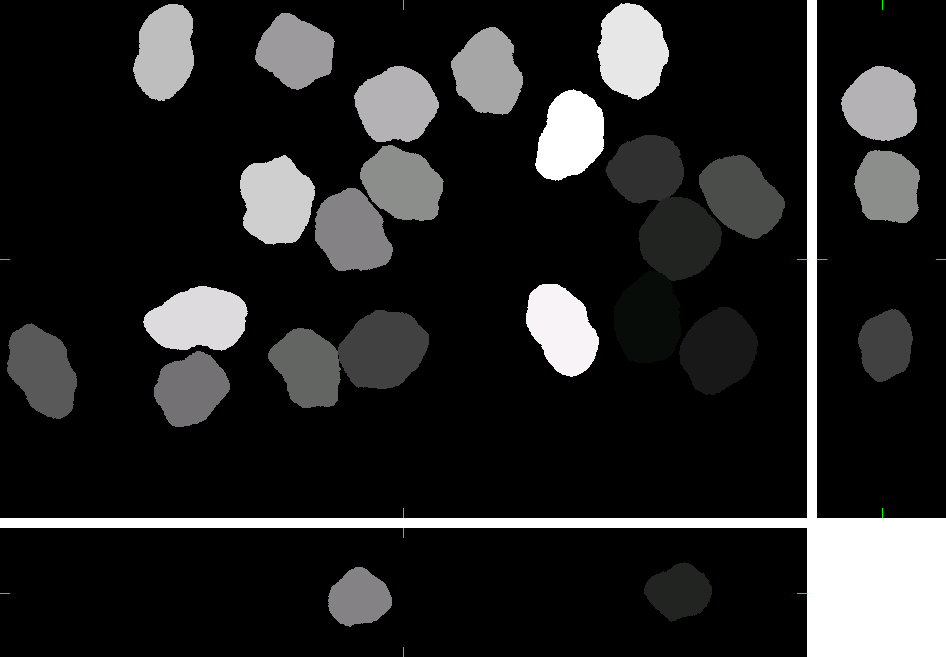
HL60 Cell Line (fixed cells)
Here, you can find 30 synthetic images of nuclei of HL60 cell line including ground truth (foreground/background) images. Each image set contains 20 cell nuclei with specified probability of clustering (0%, 25%, 50%, and 75%). The dataset was generated using the virtual microscope imitating the microscope Zeiss S100 (objective Zeiss 63x/1.40 Oil DIC) attached to confocal unit Atto CARV and CCD camera Micromax 1300-YHS. The image data was saved using three different file formats: ICS, HDF5 and 3D-TIFF. Please, feel free to select the format you prefer. All of them contain the same data.
Example images:
 |
 |
|
| 3D image | 3D labeled image |
The individual image files are aggregated in ZIP archives.
- high SNR:
- low SNR:
If you use this dataset in your research papers, please refer the following article:
- Svoboda D., Kozubek M., Stejskal S. Generation of Digital Phantoms of Cell Nuclei and Simulation of Image Formation in 3D Image Cytometry, Cytometry Part A, Volume 75A, Issue 6, pp 494-509, June 2009, ISSN:1552-4922
This research was supported by the Ministry of Education of the Czech Republic (Projects No. LC535 and No. 2B06052).
Granulocytes (fixed cells)
Here, you can find 30 synthetic images of nuclei of granulocytes including ground truth (foreground/background) images. Each image set contains up to 15 cell nuclei. The dataset was generated using the virtual microscope imitating the microscope Zeiss S100 (objective Zeiss 63x/1.40 Oil DIC) attached to confocal unit Atto CARV and CCD camera Micromax 1300-YHS. The image data was saved using three different file formats: ICS, HDF5 and 3D-TIFF. Please, feel free to select the format you prefer. All of them contain the same data. The individual image files are aggregated in ZIP archives.
Example images:
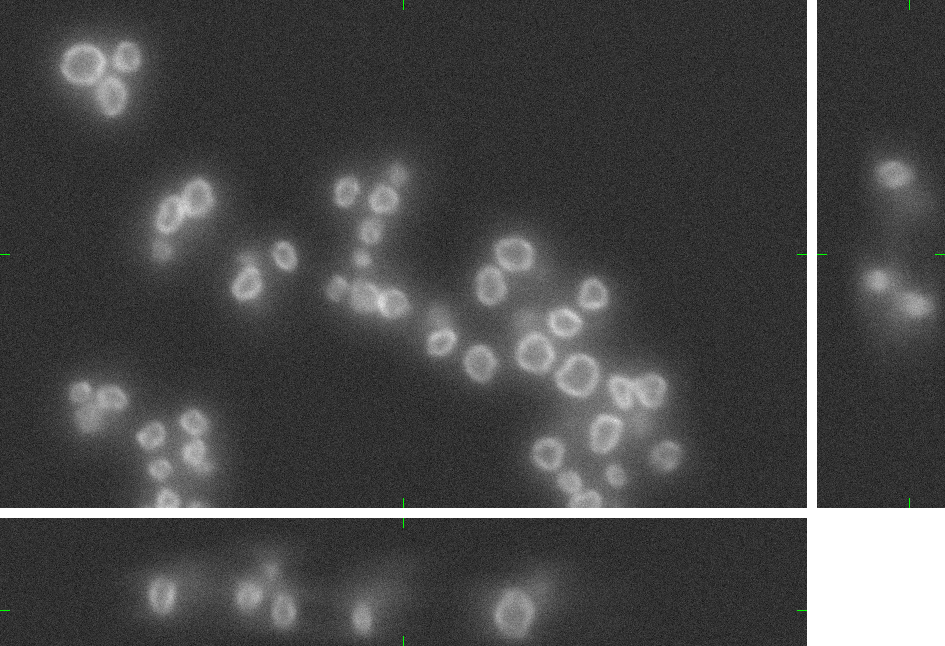 |
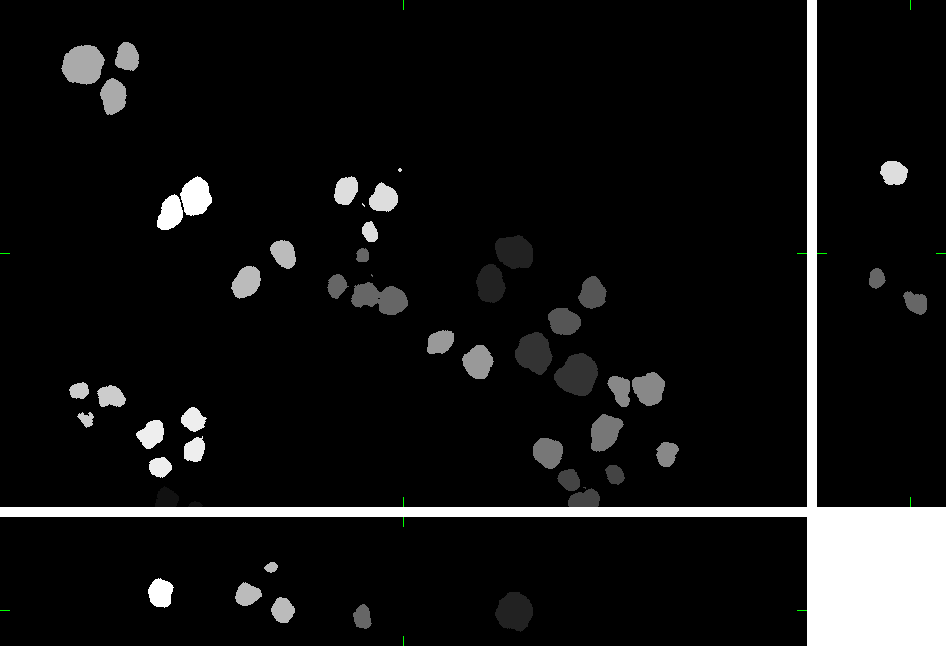 |
|
| 3D image | 3D labeled image |
If you use this dataset in your research papers, please refer the following article:
- Svoboda D., Kozubek M., Stejskal S. Generation of Digital Phantoms of Cell Nuclei and Simulation of Image Formation in 3D Image Cytometry, Cytometry Part A, Volume 75A, Issue 6, pp 494-509, June 2009, ISSN:1552-4922
This research was supported by the Ministry of Education of the Czech Republic (Projects No. LC535 and No. 2B06052).
### Time-lapse Image Sequences (living cells)
In this section, you can find a reference to collection of computer generated time-lapse image sequences of nuclei of HL60 cells stained with Hoescht (both 2D & 3D). The dataset was generated with the help of our virtual microscope. The dataset was prepared for the 1st a the 2nd edition of Cell Tracking Challenge joined to IEEE International Symposium on Biomedical Imaging
Example image sequence:
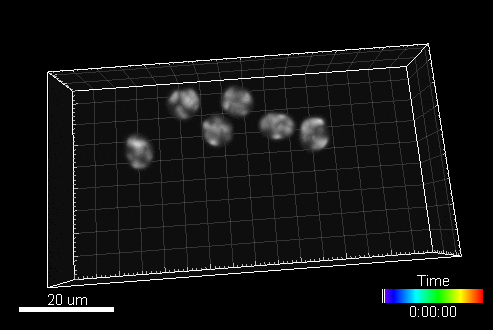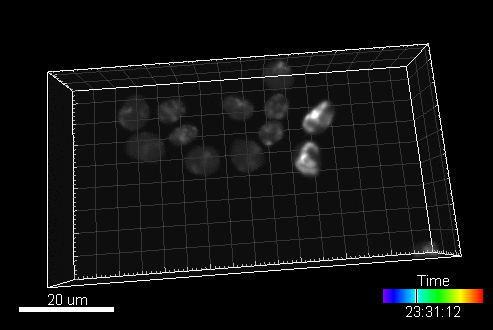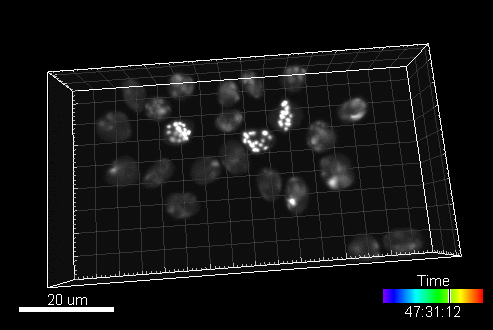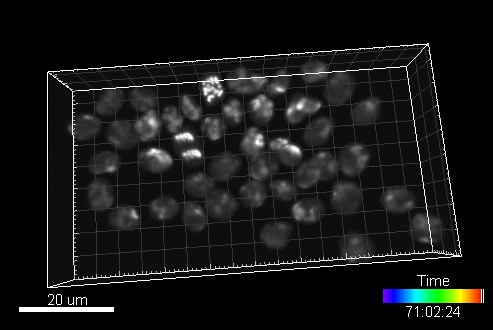
The data is available for free at this address: https://celltrackingchallenge.net/3d-datasets
If you use this dataset in your research papers, please refer one of the following articles:
-
Svoboda, D. Ulman V. MitoGen: A Framework for Generating 3D Synthetic Time-Lapse Sequences of Cell Populations in Fluorescence Microscopy. IEEE Transactions on Medical Imaging, IEEE Engineering in Medicine and Biology Society, Volume 36, Issue 1, Jan 2017, available on-line: PDF
-
Maška M, Ulman V, Svoboda D, Matula P, Matula P, Ederra C, Urbiola A, España T, Venkatesan S, Balak DM, Karas P, Bolcková; T, Štreitova M, Carthel C, Coraluppi S, Harder N, Rohr K, Magnusson KE, Jaldén J, Blau HM, Dzyubachyk O, Křížek P, Hagen GM, Pastor-Escuredo D, Jimenez-Carretero D, Ledesma-Carbayo MJ, Munoz-Barrutia A, Meijering E, Kozubek M, Ortiz-de-Solorzano C. A benchmark for comparison of cell tracking algorithms, Bioinformatics, Volume 30(11), pp 1609-1617, June 2014
This research was supported by the Czech Science Foundation, Grant No. GA14-22461S.
### Vasculogenesis (living cells)
Here, you can find the sequence of frames recording the process called vasculogenesis. The endothelial cells, initially spread across the glass slide, tend to attach each other and form the networks with thin and elongated chords. This process forces some cells to be markedly elongated, as can be seen in the figure below. The dataset was generated with the help of our virtual microscope.
Example images:
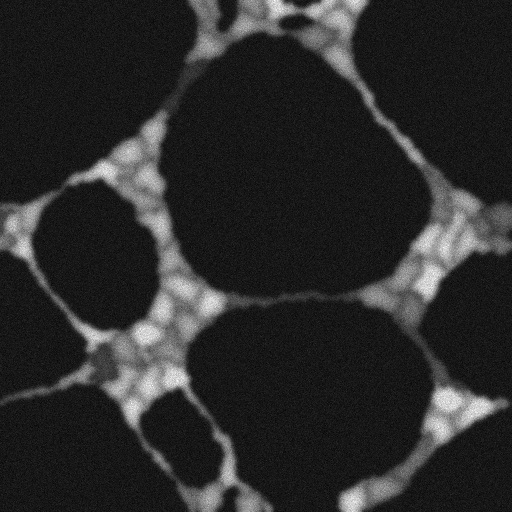 |
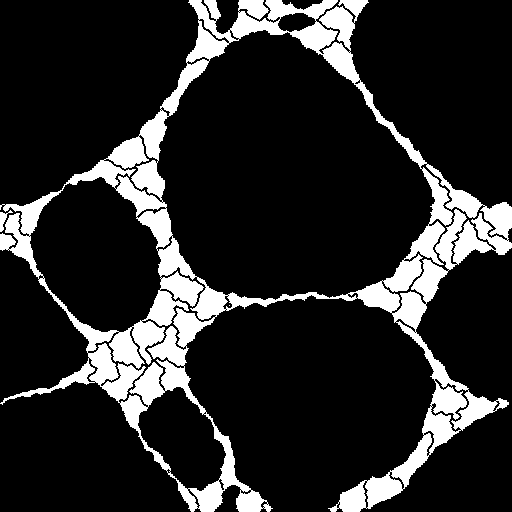 |
|
| 3D image | 3D mask |
-
sequence of images with simulated fluorescence: Zipped PNG (preview video)
-
sequence of corresponding cell labels: Zipped PNG (preview video)
If you use this dataset in your research papers, please refer the following article:
- Svoboda D, Ulman V, Kováč P, Šalingová; B, Tesařová; L, Koutná; IK, Matula P. Vascular Network Formation in Silico Using the Extended Cellular Potts Model, In IEEE International Conference on Image Processing, Phoenix, Arizona (USA), 2016, pp. 3180-3183, ISBN 978-1-4673-9961-6.
This research was supported by the Czech Science Foundation, Grant No. GA14-22461S.
### FiloData3D - Cells with filopodial protrusions (living cells)
This synthetic image data displays A549 lung cancer cells of three morphologically distinct phenotypes: wild-type (WT), CRMP-2-overexpressing (OE), and CRMP-2-phospho-defective (PD). Whereas the WT cells sporadically protrude a few single-branch filopodia that may appear and disappear, the OE cells protrude many of them with short lifetimes. By contrast, the PD cells protrude aberrantly long, branching filopodia with whole-experiment lifetimes. The image set consists of 180 synthetic 3D time-lapse sequences of single A549 lung cancer cells. The dataset includes three sequences for each of the three phenotypes, generated in 20 alternatives with five different signal-to-noise ratios and four different anisotropy ratios. Each sequence, being composed of 30 frames, was generated using FiloGen that imitates transient cell transfection with the green fluorescent protein conjugated to actin and the image formation process using a spinning disk confocal microscope equipped with a water Plan-Apochromatic 63x/1.20 objective lens.
Example images:
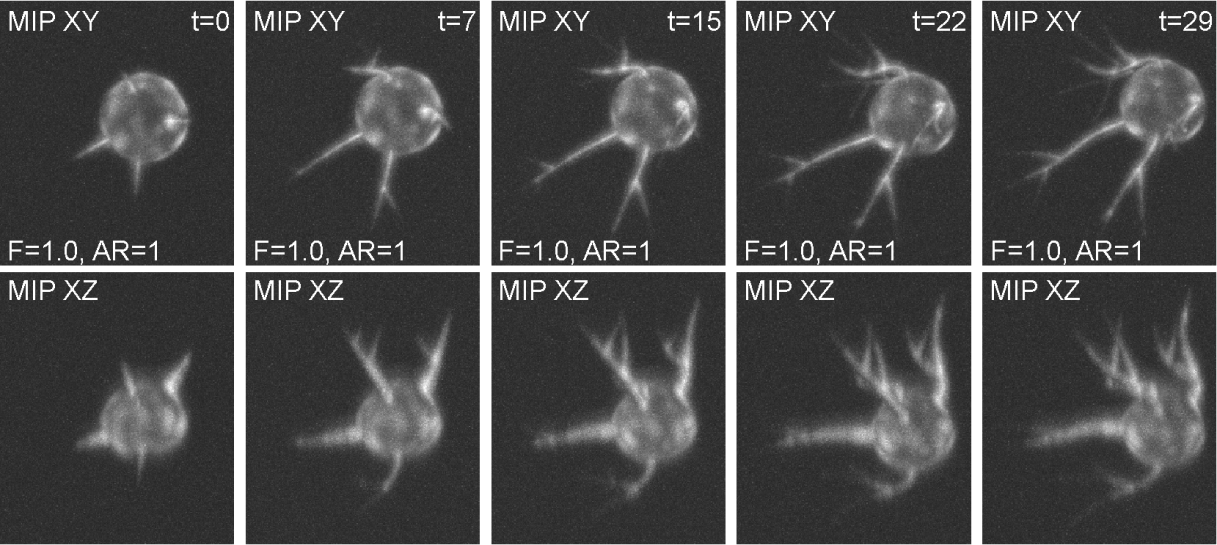
The synthetic time-lapse image data of a CRMP-2-phospho-defective cell for the given fluorescence level factor (F=1.0) and anisotropy ratio (AR=1).
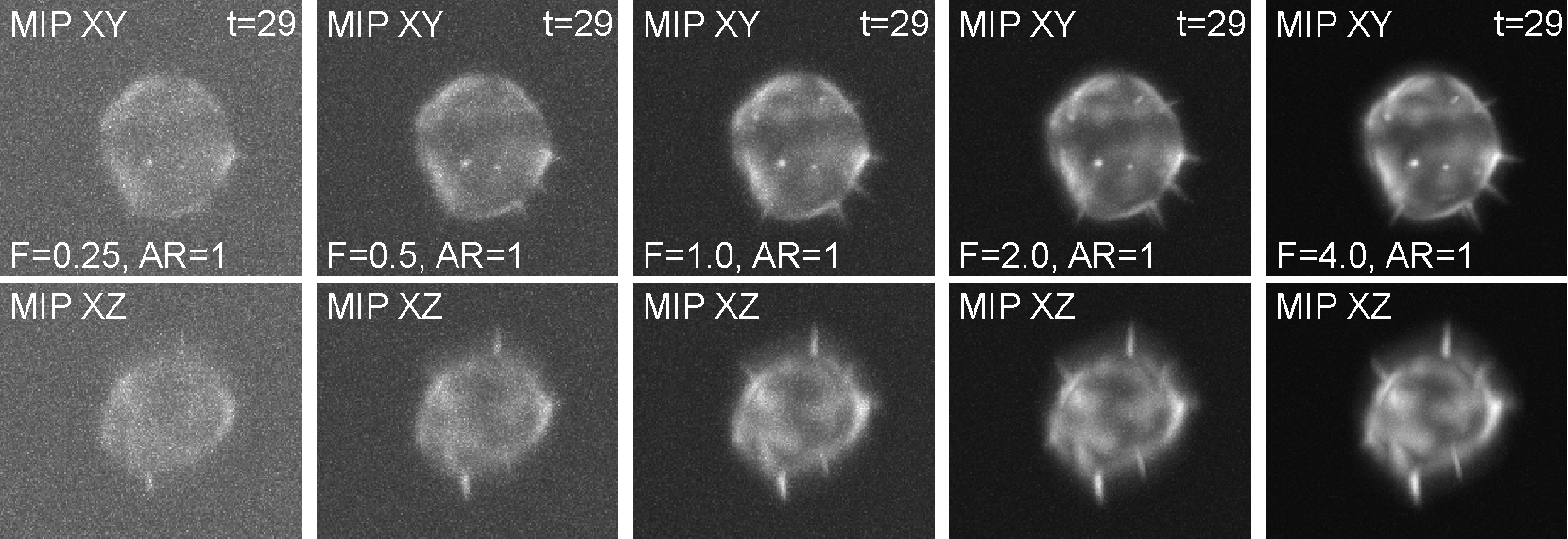
The synthetic image data of a CRMP-2-overexpressing cell for varying fluorescence level factors (Fs), which indirectly affect the signal-to-noise ratios of the image data.
If you use this dataset, being tagged as BBBC046 in the Broad Bioimage Benchmark Collection, in your research papers, please refer to the following articles:
- Dmitry V. Sorokin, Igor Peterlík, Vladimír Ulman, David Svoboda, Tereza Nečasová, Katsiarina Morgaenko, Lívia Eiselleová, Lenka Tesařová, Martin Maška. FiloGen: A Model-Based Generator of Synthetic 3-D Time-Lapse Sequences of Single Motile Cells with Growing and Branching Filopodia. IEEE Transactions on Medical Imaging, 37(12):2630-2641, 2018.
- Martin Maška, Tereza Nečasová, David Wiesner, Dmitry V. Sorokin, Igor Peterlík, Vladimír Ulman, David Svoboda. Toward Robust Fully 3D Filopodium Segmentation and Tracking in Time-Lapse Fluorescence Microscopy. In IEEE International Conference on Image Processing, 2019, pp. 819-823.
This work was supported by the Czech Science Foundation under the grants number GJ16-03909Y and GA17-05048S, the Czech Ministry of Education, Youth and Sports under the grants number LTC17016 and CZ.02.1.01/0.0/0.0/16_013/0001775, the Russian Science Foundation under the grant number 17-11-01279, and the German Federal Ministry of Research and Education (BMBF) under the grant number 031L0102 (de.NBI).
Full segmentation annotations of 3D time-lapse microscopy images of MDA231 cells
In the field of biomedical image analysis, one of the richest benchmark datasets for objective comparison of cell segmentation and tracking methods is the Cell Tracking Challenge (CTC). The CTC offers two types of 3D reference annotations: Gold reference tracking annotations (TRA) and Silver reference segmentation annotations (ST).
We present fully volumetric manual annotations for two temporal sequences of a 3D CTC dataset, Fluo-C3DL-MDA231, which is characterized by the most complex cell shapes out of all 3D CTC datasets.
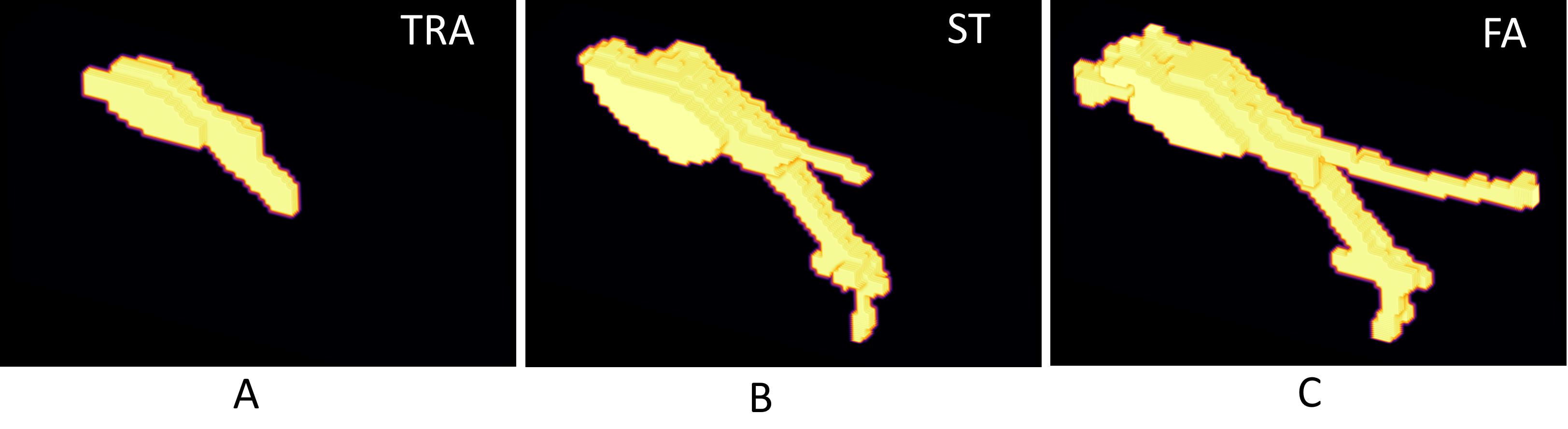
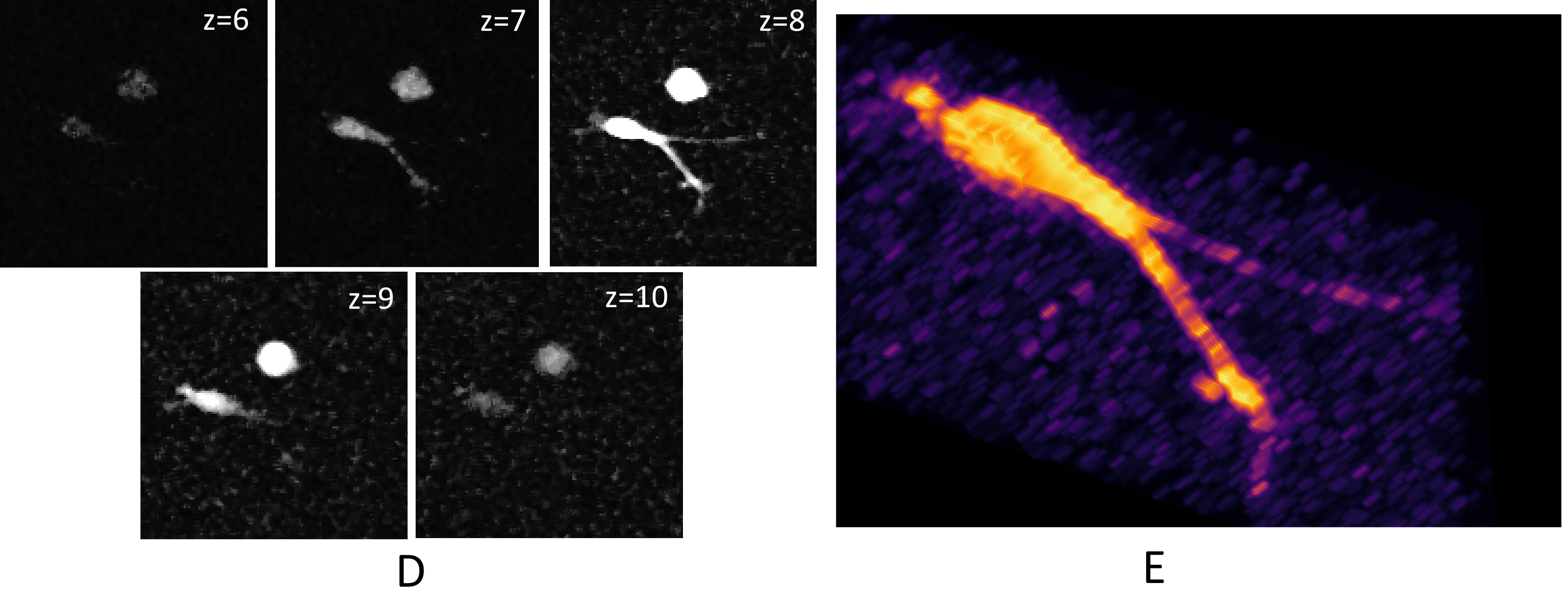
A 3D visualization of one cell mask that consists of 5 slices (w.r.t. the z-axis). The first row shows a 3D visualization of the 3 annotations: (A) Gold reference tracking annotations (TRA), (B) Silver reference segmentation annotations (ST), and (C) the proposed full annotations (FA). Image (D) shows all five 2D slices of the cell. Image (D) shows a 3D visualization of the slices (D). It is visible that thin protrusions are absent in images (A) and (B).
There are 3 annotations available (A1, A2, A3) for the first sequence (S01), and one annotation (A1) is available for the second sequence (S02). The annotations A1, A2, and A3 were merged by a simple majority-voting procedure to acquire the consensus annotation for sequence S01. The majority-voting (MV) procedure implies that all pixels having less than 2 votes for the same cell mask were removed from the final annotation. All annotations were saved as multi-layered unit16 gray-scale tif images
The respective microscopy images can be dowloaded from CTC website: https://celltrackingchallenge.net/3d-datasets/
We provide three human annotations for the first sequence and one for the second. For the first sequence, we also provide a result of majority voting.
The full annotations are consistent with CTC tracking annotations (TRA) and will be kept up-to-date in the future. Soon, we plan to provide more annotations and majority voting for the second sequence.
The dataset is used in a paper submitted to the ISBI 2025 conference, and further details are available in the corresponding arXiv publication:
- Melnikova, A. and Matula, P., 2025, April. Study of Shape Fusion Algorithms for 3D Time-Lapse Microscopy. In 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI) (pp. 1-5). IEEE.
- Melnikova, A. and Matula, P., 2025. Full segmentation annotations of 3D time-lapse microscopy images of MDA231 cells. arXiv preprint arXiv:2510.10797.
The work was founded by the Czech Science Foundation, project no. GA21-20374S.