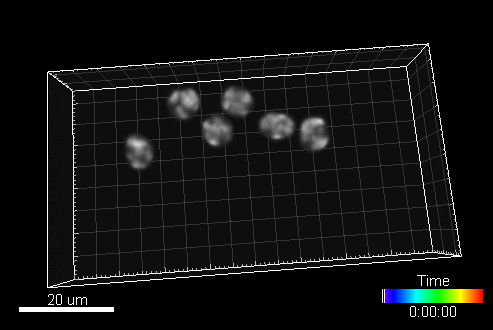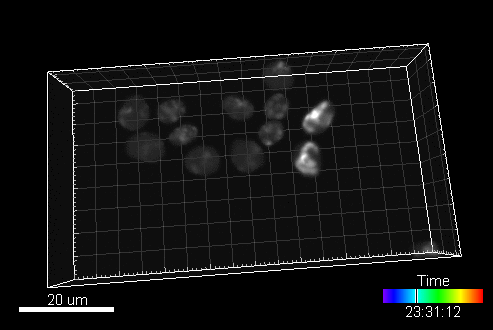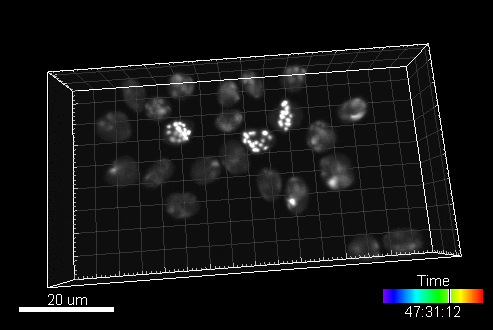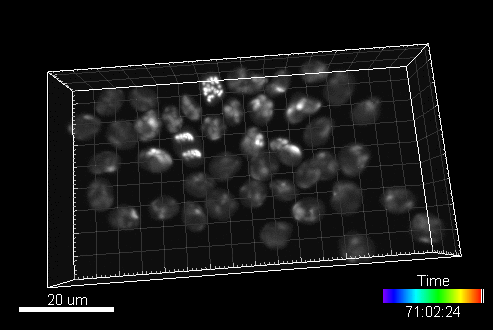
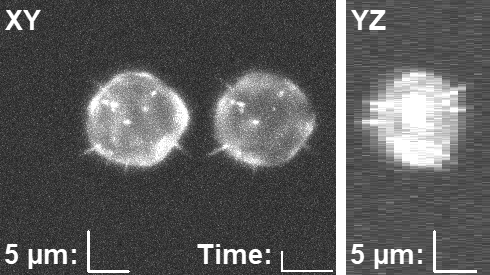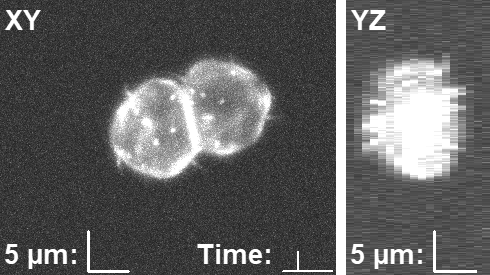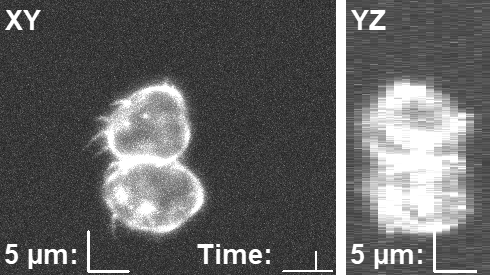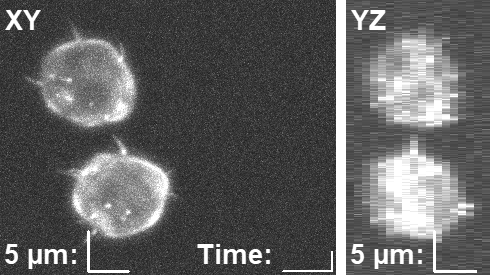
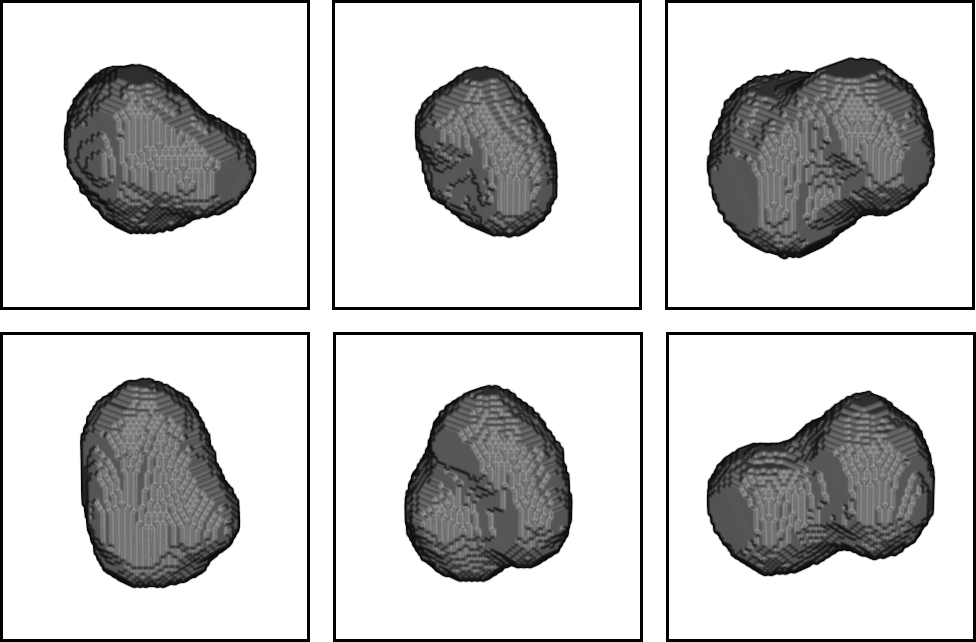
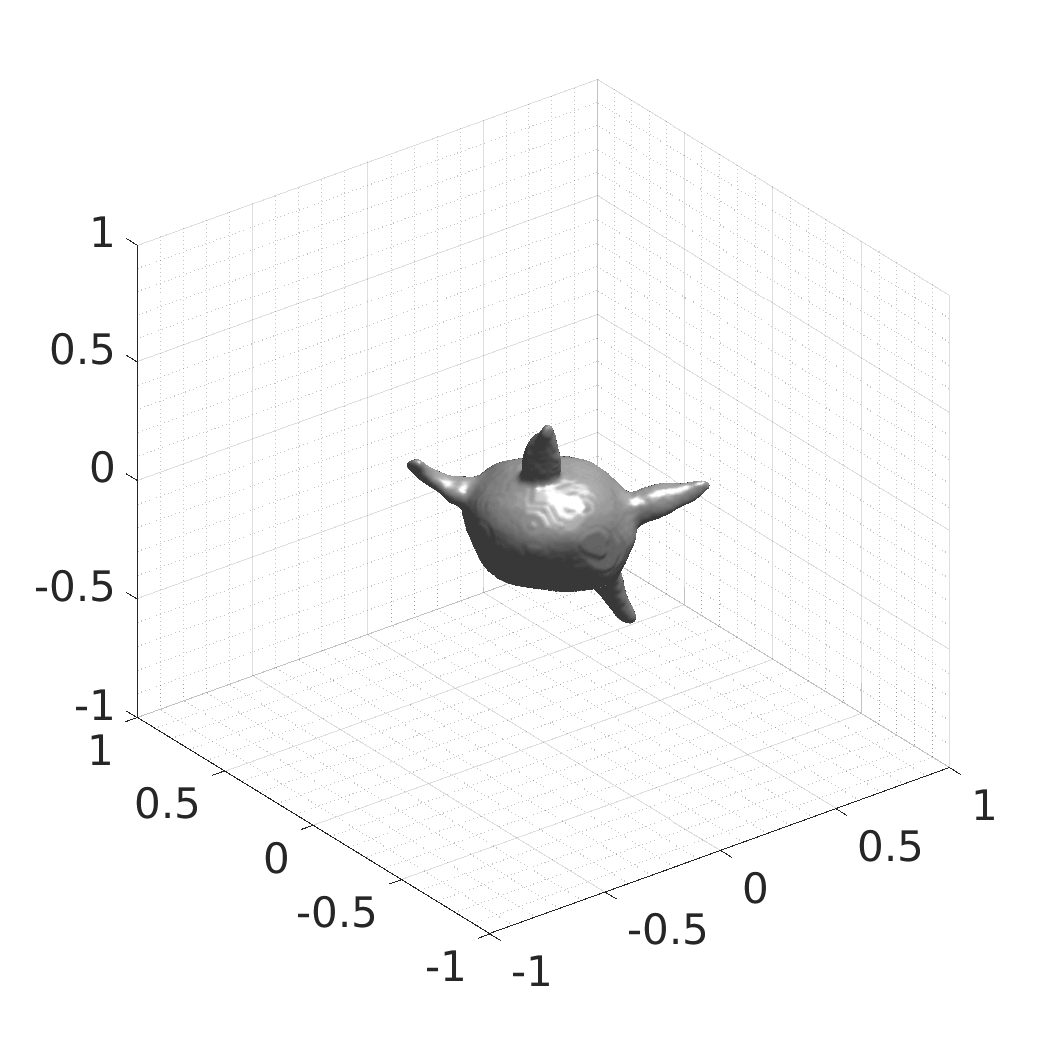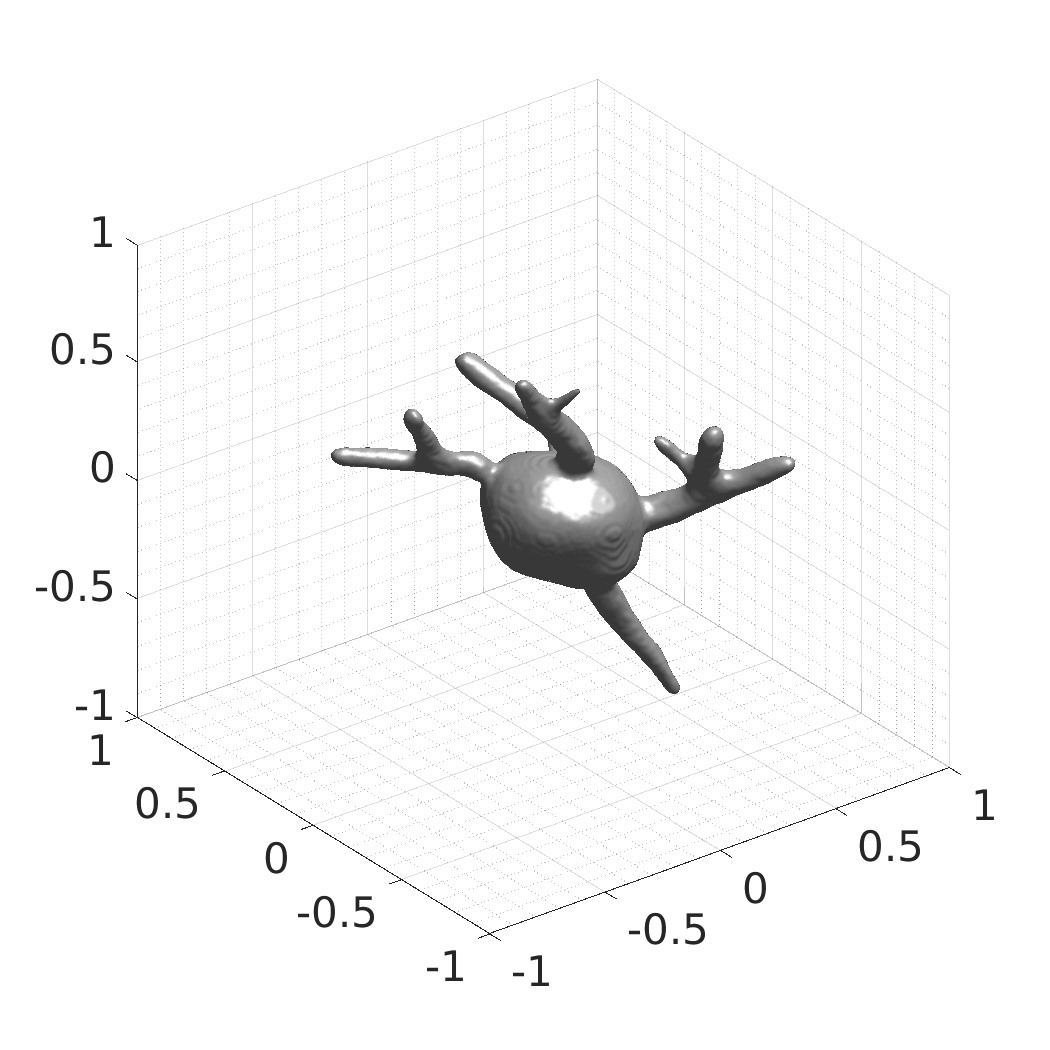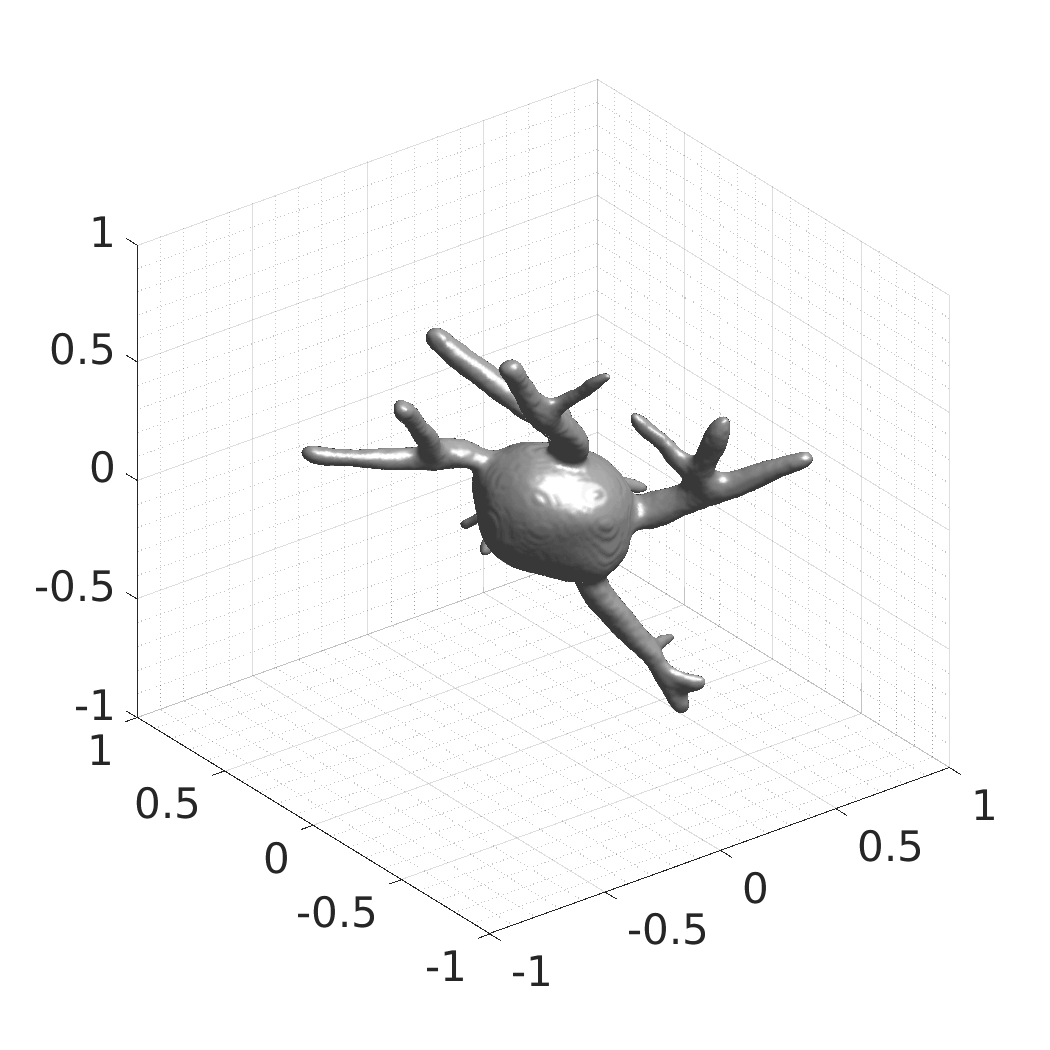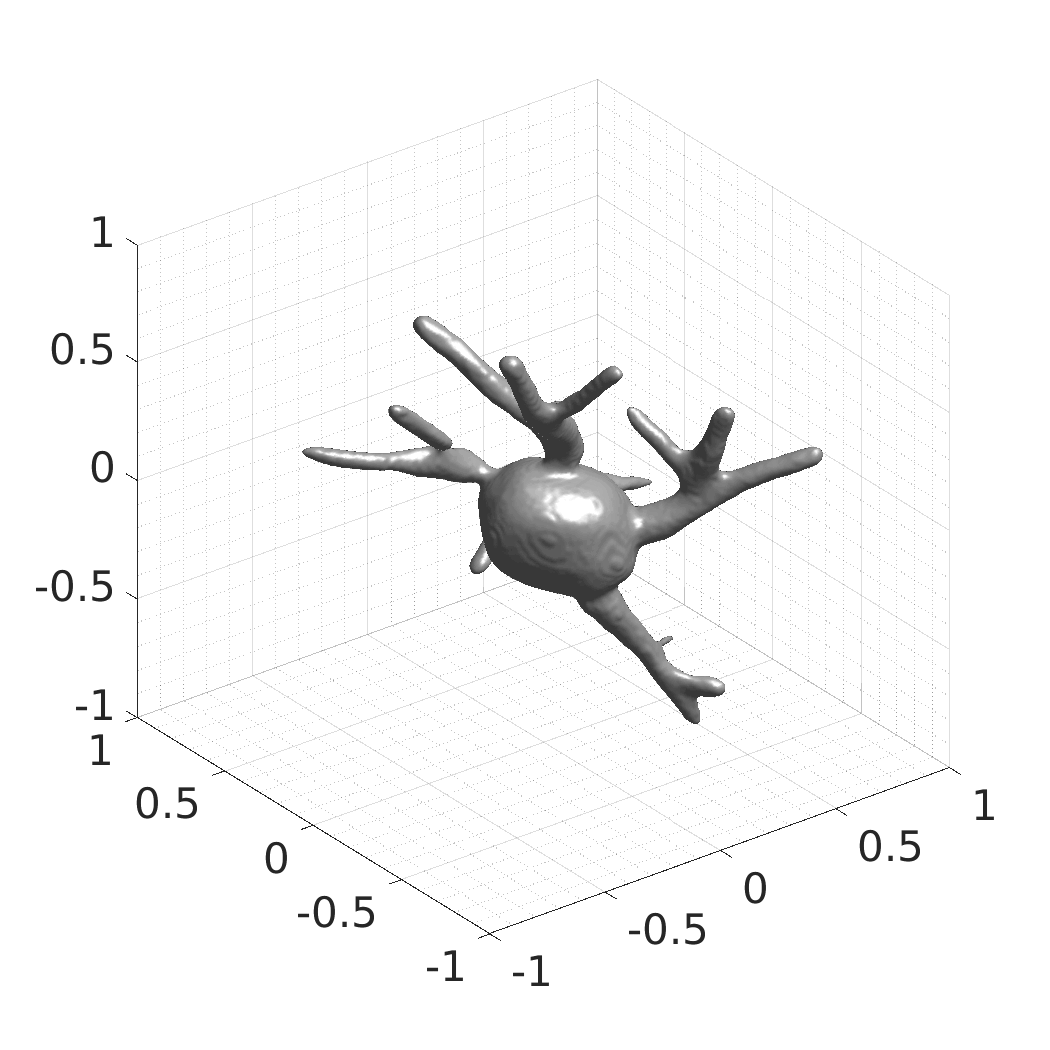
CytoPacq
A web-based simulation framework capable of generating artificial microscopy image data fully in 3D+time. The simulated data imitate the image as if acquired using the fluorescence optical microscope.
Feel free to check this service and use the generated data!