CytoGen
CytoGen project aims to generate four types of digital phantoms: HL-60 cell nuclei, granulocyte nuclei, microspheres, and colon tissues. The generated digital phantoms are characterized both with its shape and an internal structure. CytoGen serves as a module in CytoPacq framework.
Sample data
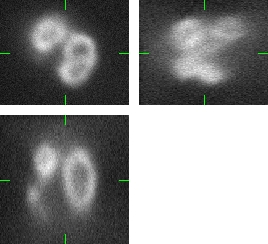
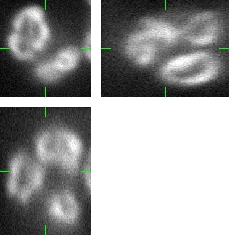
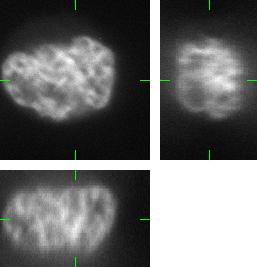
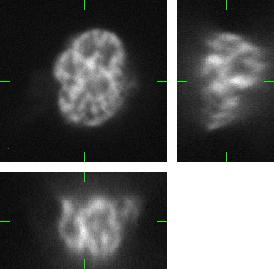
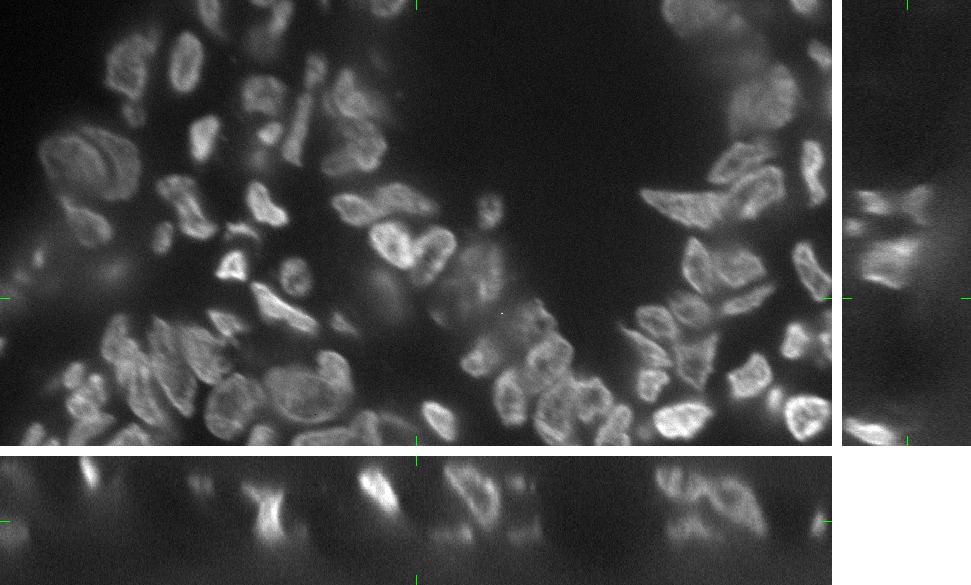
The generated image data are fully 3D images. For visualization purposes we use three orthogonal projections: xy, xz, and yz. Comparing the Fig.1 and Fig.2, you can observe the plausability of the synthetic data.
 (a) |
 (b) |
 (c) |
 (d) |
 (e) |
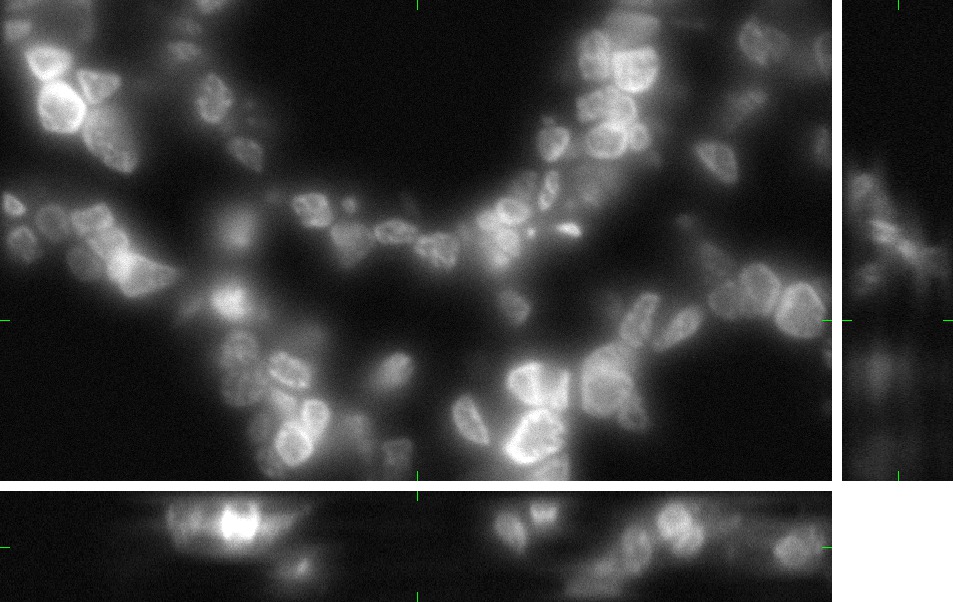
Fig 1.: An example of five real images of (a-b) two granulocyte nuclei, (c-d) two HL60 nuclei and (e) one tissue colon section. Each 3D figure consists of three individual images: the top-left image contains selected xy-slice, the top-right image corresponds to selected yz-slice, and the bottom one depicts selected xz-slice. Three mutually orthogonal slice planes are shown with ticks.
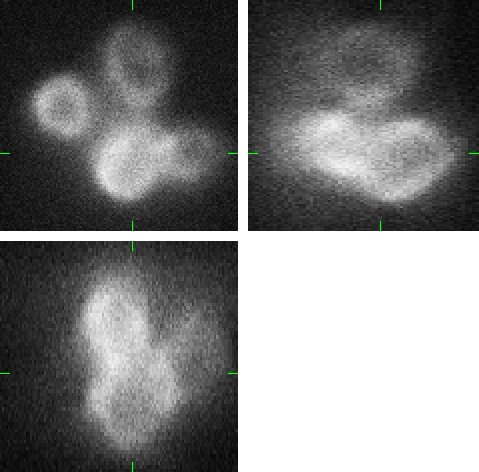 (a) |
 (b) |
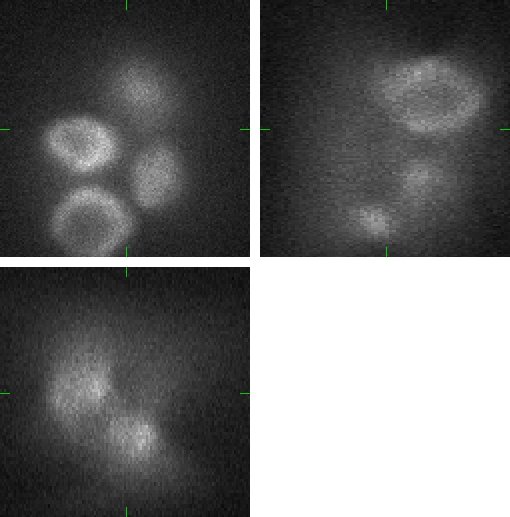
 (c) |
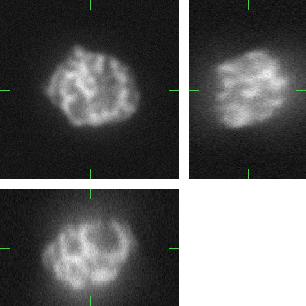
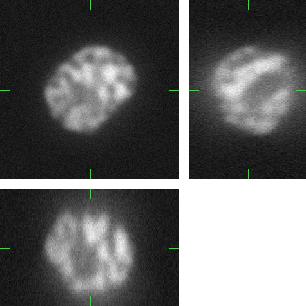 (d) |
 (e) |
Fig 2.: An example of five synthetic images of (a-b) two granulocyte nuclei, (c-d) two HL60 nuclei and (e) one tissue colon section. Each 3D figure consists of three individual images: the top-left image contains selected xy-slice, the top-right image corresponds to selected yz-slice, and the bottom one depicts selected xz-slice. Three mutually orthogonal slice planes are shown with ticks.
Download
The source codes are freely available and are subject to GNU GPL. Before you start compiling, building or running this application, please read the documentation carefully.
In order to prevent the potential users from being annoyed when downloading all the necessary libraries and compiling them together with the source codes of this project, we prepared a virtual computer with all the libraries and CytoGen built and ready for an immediate use. The virtual computer has been prepared in Virtual Box environment (version 6) and is available here (5.3 GB). The installed operating system that hosts CytoGen is Linux Ubuntu OS with Xfce environment. After downloading and running the virtual computer you will be requested to enter the password. Please, type “cbia” and enjoy using CytoGen! If you meet any difficulties or obstacles during running CytoGen, please feel free to contact us. We appreciate your feedback and will be happy to assist you.
References
-
David Svoboda, Vladimír Ulman. Towards a Realistic Distribution of Cells in Synthetically Generated 3D Cell Populations. In Alfredo Petrosino. 17th International Conference on Image Analysis and Processing - ICIAP 2013. LNCS 8157, Part II. Berlin, Heidelberg: Springer-Verlag, 2013. p. 429-438, 10 pp. ISBN 978-3-642-41183-0.
-
David Svoboda. Efficient Computation of Convolution of Huge Images. In Giuseppe Maino; Gian Luca Foresti. Image Analysis and Processing - ICIAP 2011. LNCS 6978, Part I. Berlin, Heidelberg: Springer-Verlag, 2011. p. 453-462, 10 pp. ISBN 978-3-642-24084-3.
-
David Svoboda, Ondřej Homola, Stanislav Stejskal. Generation of 3D Digital Phantoms of Colon Tissue. In M. Kamel and A. Campilho. Proceedings of 8th International Conference on Image Analysis and Recognition. LNCS 6754, Part II. Berlin, Heidelberg: Springer-Verlag, 2011. p. 31-39, 9 s. ISBN 978-3-642-21595-7.
-
David Svoboda, Michal Kozubek, Stanislav Stejskal. Generation of Digital Phantoms of Cell Nuclei and Simulation of Image Formation in 3D Image Cytometry. Cytometry Part A, John Wiley & Sons, Inc., 2009, 75A, No 6, p. 494-509. ISSN 1552-4922.
-
David Svoboda, Marek Kašík, Martin Maška, Jan Hubený, Stanislav Stejskal and Michal Zimmermann. On Simulating 3D Fluorescent Microscope Images. In Computer Analysis of Images and Patterns. Berlin, Heidelberg: Springer-Verlag, 2007. p. 309-316, 8 pp. ISBN 978-3-540-74271-5.
Acknowledgement
The progress of work devoted to the simulator was incremental in order to meet actual needs of scientific projects No. 2B06052 and No. LC535 by the Ministry of Education of the Czech Republic and the running scientific projects GBP302/12/G157 and GA14-22461S by the Czech Science Foundation.