FiloGen
FiloGen is a model-based generator of synthetic 3D time-lapse sequences of single motile cells with growing and branching filopodial protrusions. It allows one to generate realistic, fully annotated 3D+t image data for objective benchmarking of filopodium segmentation and tracking approaches.
Summary
The existence of diverse image datasets accompanied by reference annotations is a crucial prerequisite for an objective benchmarking of bioimage analysis methods. Nevertheless, such a prerequisite is arduous to satisfy for time-lapse, multidimensional fluorescence microscopy image data, manual annotations of which are laborious and often impracticable. FiloGen is a simulation system capable of generating 3D time-lapse sequences of single motile cells with filopodial protrusions of user-controlled structural and temporal attributes, such as the number, thickness, length, level of branching, and lifetime of filopodia, accompanied by inherently generated reference annotations. The simulation system involves three globally synchronized modules, each being responsible for a separate task: the evolution of filopodia on a molecular level, linear elastic deformation of the entire cell with filopodia, and the synthesis of realistic, time-coherent cell texture. Its flexibility has been demonstrated by generating multiple synthetic 3D time-lapse sequences of single actin-stained A549 lung cancer cells stably transfected with two genetically different variants of the CRMP-2 protein, qualitatively and quantitatively resembling their real counterparts acquired using a confocal fluorescence microscope.
Preview of Sample Image Data
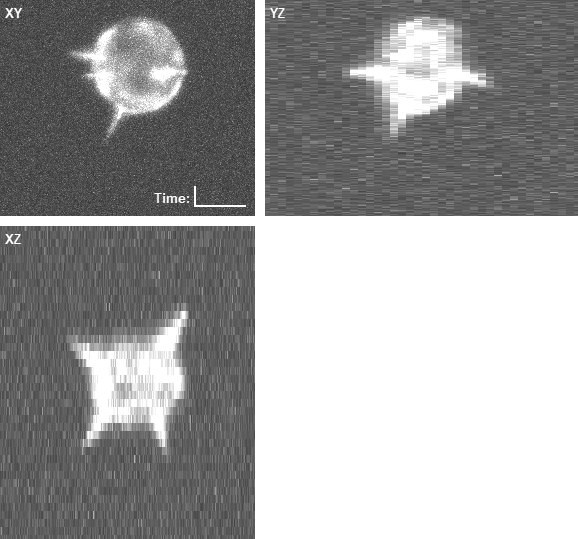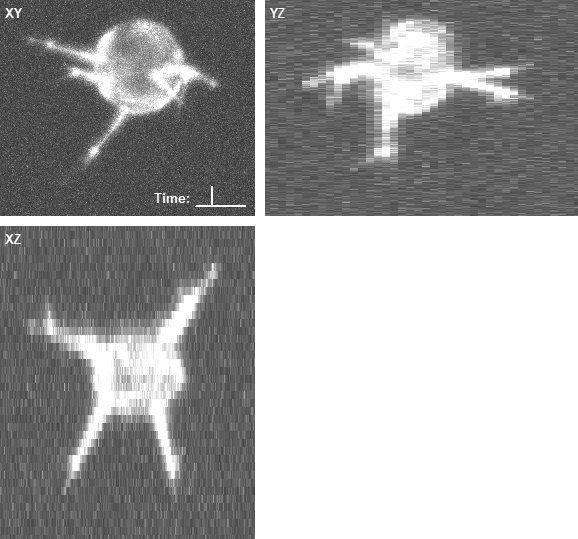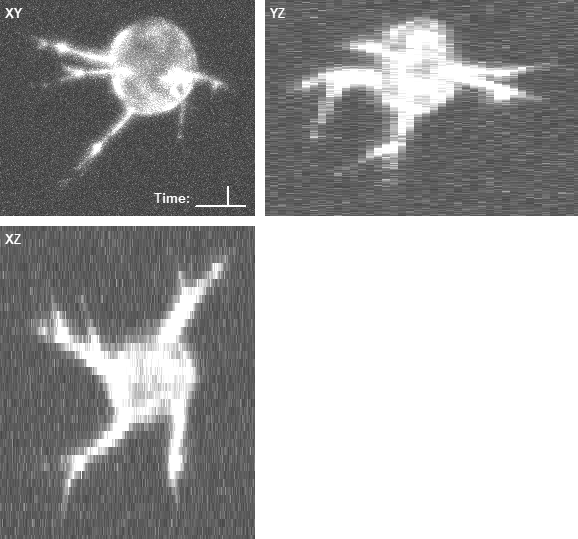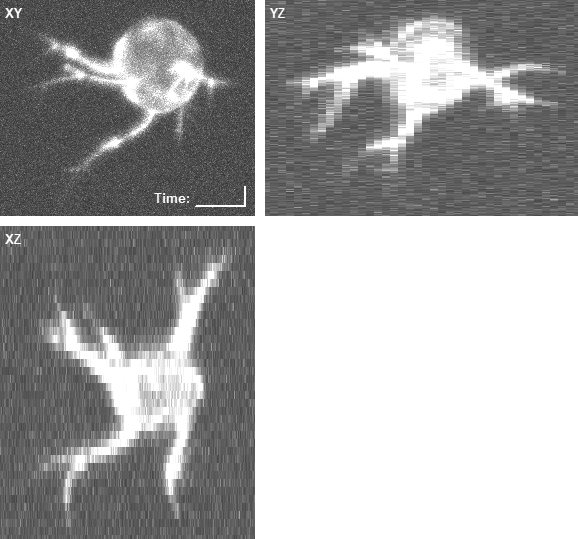
In both cases, the three panels in the right animations display the maximum intensity projections along the z (XY), x (YZ), and y (XZ) axes, with the intensities enhanced by a percentile stretch for better visibility. The voxel size is 0.126 × 0.126 × 1.0 microns.
- An example of the rendered geometry and the corresponding image data of a computer-generated overexpressing cell:


- An example of the rendered geometry and the corresponding image data of a computer-generated phospho-defective cell:

Resources
The computer-generated datasets, each consisting of three sequences, of overexpressing and phospho-defective lung cancer cells, along with their complete segmentation and tracking annotations, can be downloaded here.
The FiloGen modules were designed as separate software components that exchange their data by reading from and writing to files. The geometry module was implemented in Matlab R2017a. The deformation and texture modules were implemented in C++, the former being built on the SOFA framework. The three modules, including their supervisor and the source codes, were integrated into a pre-configured virtual machine (VM; approximately 6.6 GB to download and 18 GB to extract). A detailed description of the VM content can be found in the filogen-readme.txt file, available on the VM desktop.
Conditions of Use
The resources provided are free of charge for noncommercial and research purposes. Their use for any other purpose is generally possible, but solely with the explicit permission of the authors. In case of any questions, please do not hesitate to contact us at xmaska@fi.muni.cz.
In addition, you are expected to include adequate references whenever you present or publish results that are based on the resources provided.
References
- Dmitry V. Sorokin, Igor Peterlík, Vladimír Ulman, David Svoboda, Tereza Nečasová, Katsiarina Morgaenko, Lívia Eiselleová, Lenka Tesařová, Martin Maška. FiloGen: A Model-Based Generator of Synthetic 3-D Time-Lapse Sequences of Single Motile Cells with Growing and Branching Filopodia. IEEE Transactions on Medical Imaging, 37(12):2630-2641, 2018.
- Dmitry V. Sorokin, Igor Peterlík, Vladimír Ulman, David Svoboda, Martin Maška. Model-Based Generation of Synthetic 3D Time-Lapse Sequences of Motile Cells with Growing Filopodia. In IEEE International Symposium on Biomedical Imaging, 2017, pp. 822–826.
Acknowledgment
This work was supported by the Czech Science Foundation under the grant number GJ16-03909Y, the Russian Science Foundation under the grant number 17-11-01279, and the German Federal Ministry of Research and Education (BMBF) under the grant number 031L0102 (de.NBI).