MitoGen
The MitoGen project aims at a generation of synthetic images of living cell populations. The synthetic cells can move, split during mitosis, and enter or leave the region of interest in the virtual microscopy slide. The MitoGen project serves as a module in a CytoPacq framework which is capable of simulating the operation of the whole optical microscope (including the CCD camera). It produces time-lapse sequences of 3D volumetric image data.
Sample Data
A sample image sequence generated by MitoGen (visualized in Imaris software):
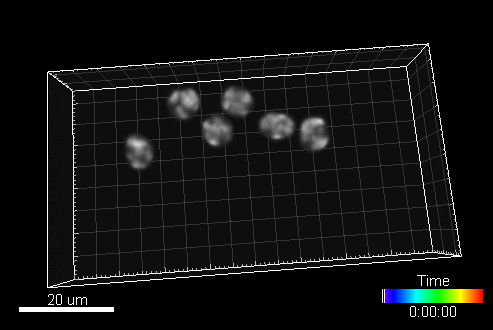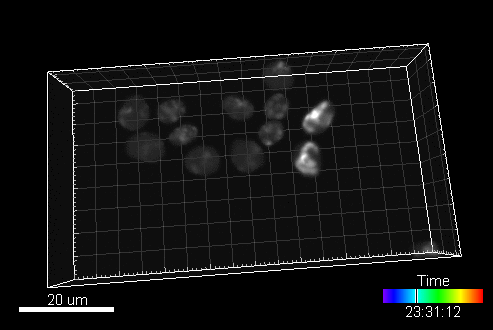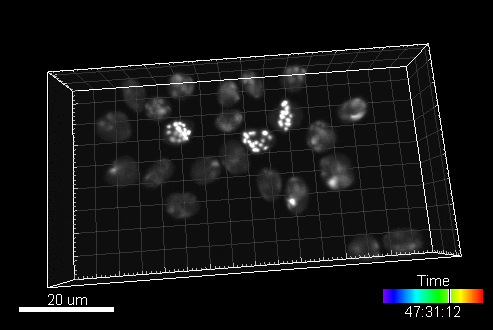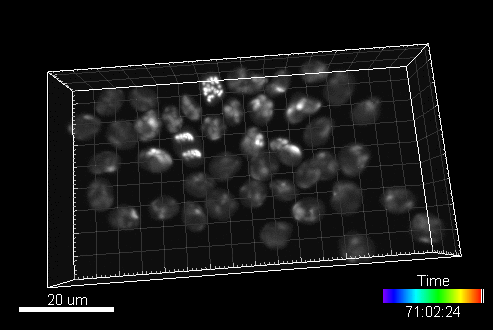
The image data (time-lapse sequences) generated by MitoGen has already been used in the 1st, 2nd, and 3rd edition of Cell Tracking Challenge joined to IEEE International Symposium on Biomedical Imaging.
Download
MitoGen is written in C++. It uses TIFF and ICS image formats for storing the generated image data. It uses GNU GSL for random number generations. Last but not least, it also uses the CBIA i3dlibs to generate the images of the cells.
Source code with CMake configuration script (developed and tested in Linux OS), and input data is available for free. In order to prevent the potential users from being annoyed when downloading all the necessary libraries and compiling them together with the source codes of this project, we prepared a virtual computer with all the libraries and MitoGen built and ready for an immediate use. The virtual computer has been prepared in Virtual Box environment (version 6) and is available here (5.3 GB). The installed operating system that hosts MitoGen is Linux Ubuntu OS with Xfce environment. After downloading and running the virtual computer you will be requested to enter the password. Please, type “cbia” and enjoy using MitoGen! If you meet any difficulties or obstacles during running MitoGen, please feel free to contact us. We appreciate your feedback and will be happy to assist you.
Note: The MitoGen is computationally intensive program. We recommend to download and run our virtual computer on a powerfull machine (at least 32 GB RAM) with the support of virtualization technology enabled in BIOS. If you use Windows OS, disable the Hyper-V feature in your system.
License
The MitoGen is freely available under the GNU GPL license. Copyright belongs to the Centre for Biomedical Image Analysis (CBIA) at Masaryk University.
References
The MitoGen project is described in detail in the following journal paper:
- David Svoboda, Vladimír Ulman. MitoGen: A Framework for Generating 3D Synthetic Time-Lapse Sequences of Cell~Populations in Fluorescence Microscopy. IEEE Transactions on Medical Imaging, 36(1):310-321, 2017.
Other papers related to this project include:
-
David Svoboda, Vladimír Ulman. Generation of Synthetic Image Datasets for Time-Lapse Fluorescence Microscopy. In Campilho, Aurélio; Kamel, Mohamed. Proceedings of 9th International Conference on Image Analysis and Recognition (ICIAR). LNCS 7325, Part II. Heidelberg: Springer-Verlag, 2012. p. 473-482, 10 p. ISBN 978-3-642-31297-7.
-
David Svoboda, Michal Kozubek, Stanislav Stejskal. Generation of Digital Phantoms of Cell Nuclei and Simulation of Image Formation in 3D Image Cytometry. Cytometry Part A, John Wiley & Sons, Inc., 2009, 75A, No 6, p. 494-509. ISSN 1552-4922.
-
Maška M, Ulman V, Svoboda D, Matula P, Matula P, Ederra C, Urbiola A, España T, Venkatesan S, Balak DM, Karas P, Bolcková T, Štreitova M, Carthel C, Coraluppi S, Harder N, Rohr K, Magnusson KE, Jaldén J, Blau HM, Dzyubachyk O, Křížek P, Hagen GM, Pastor-Escuredo D, Jimenez-Carretero D, Ledesma-Carbayo MJ, Muñoz-Barrutia A, Meijering E, Kozubek M, Ortiz-de-Solorzano C. A benchmark for comparison of cell tracking algorithms, Bioinformatics, Volume 30(11), pp 1609-1617, June 2014. ISSN 1367-4803.
Acknowledgement
This project was funded by Czech Science Foundation, grant No. GA14-22461S.