Reliable Segmentation and Tracking of Organoids
This web page supplements the paper entitled Segmentation and Tracking of Mammary Epithelial Organoids in Brightfield Microscopy, which introduces a deep-learning-based pipeline for automatic segmentation and tracking of organoids of interest in 2D+t high-resolution brightfield microscopy sequences.
Summary
Our organoid analyzer reliably segments and tracks mammary epithelial organoids of morphologically different phenotypes, manifesting itself as a reproducible and laborless alternative to manual processing of the acquired image data. It involves a four-layer U-Net to infer semantic segmentation predictions, adaptive morphological filtering to establish candidate organoid instances, and a shape-similarity-constrained, instance-segmentation-correcting tracking step to associate the corresponding organoid instances in time. The segmentation network was purposely trained using plausible computer-generated image data produced by our pix2pixHD-based organoid texture simulator. Such a semantic-information-rich synthesis helped the network learn robust spatial features to accurately delineate the single-organoid boundary even in challenging cases when out-of-focus structures collide with the organoid of interest.
Preview of Image Data
Segmentation results of our organoid analyzer for cysts (C), normally branching (NB), massively branching (MB), hollowly branching (HB), and long branching (LB) organoids, and cysts co-cultured with fibroblasts (FIB), with the white bars corresponding to 100 microns:
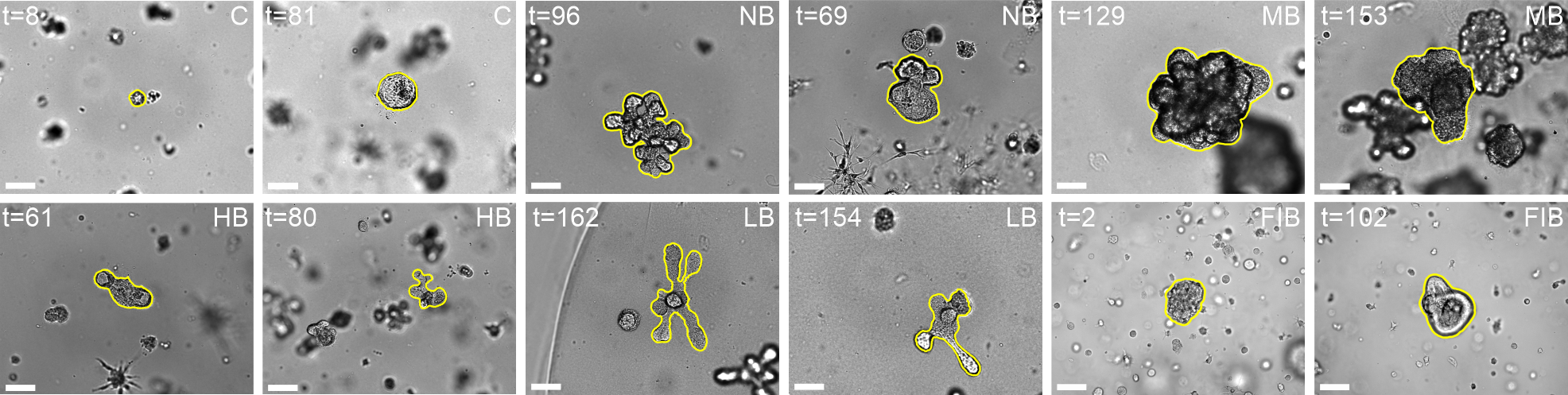
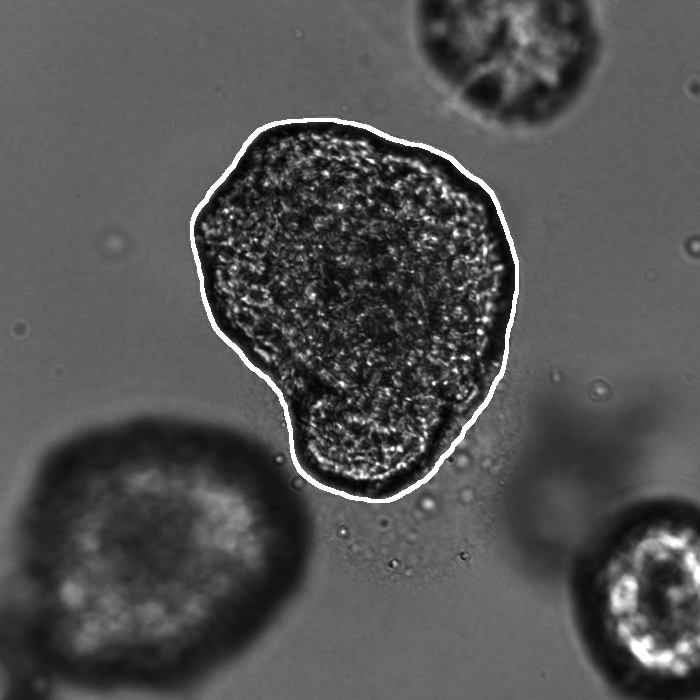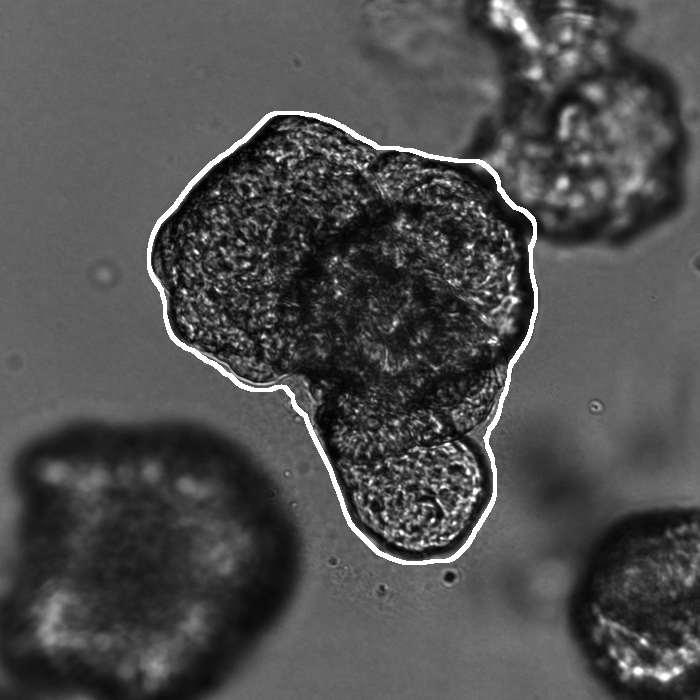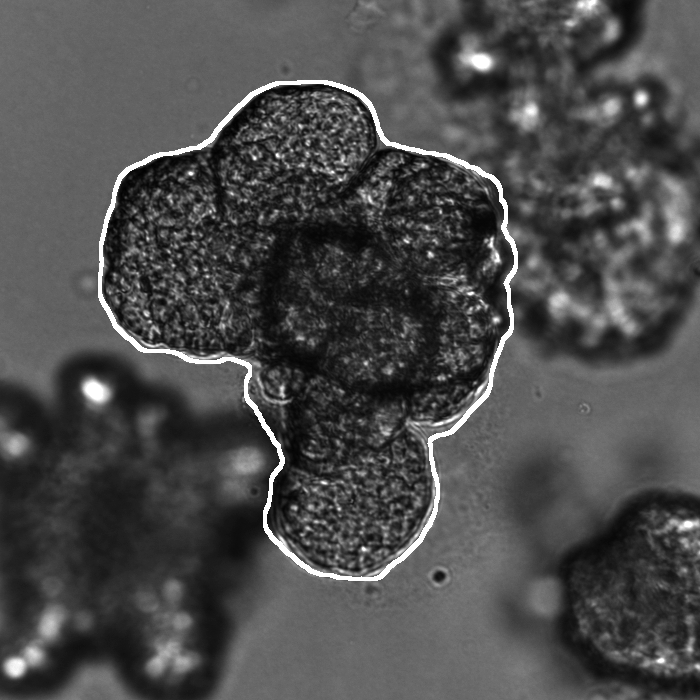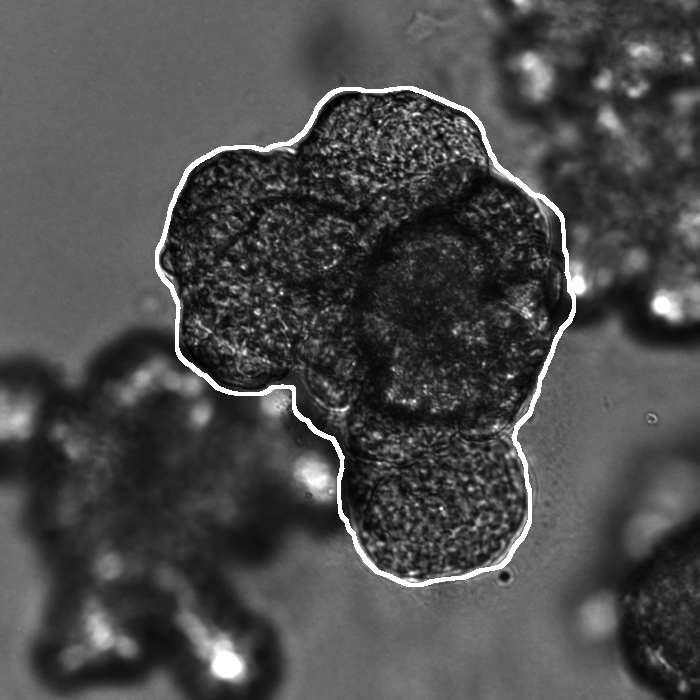
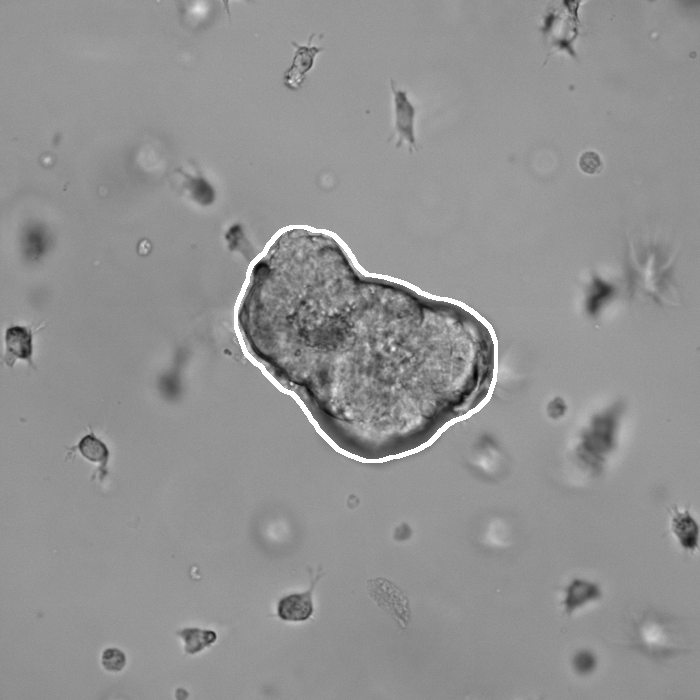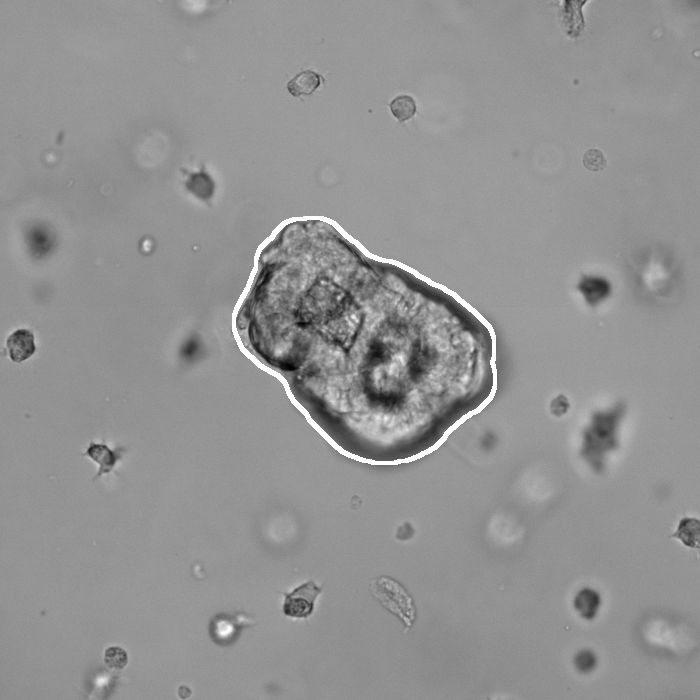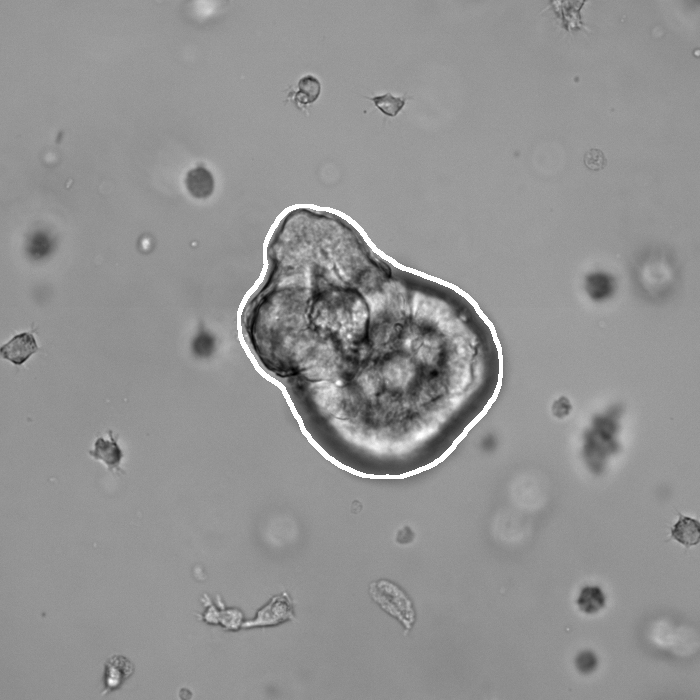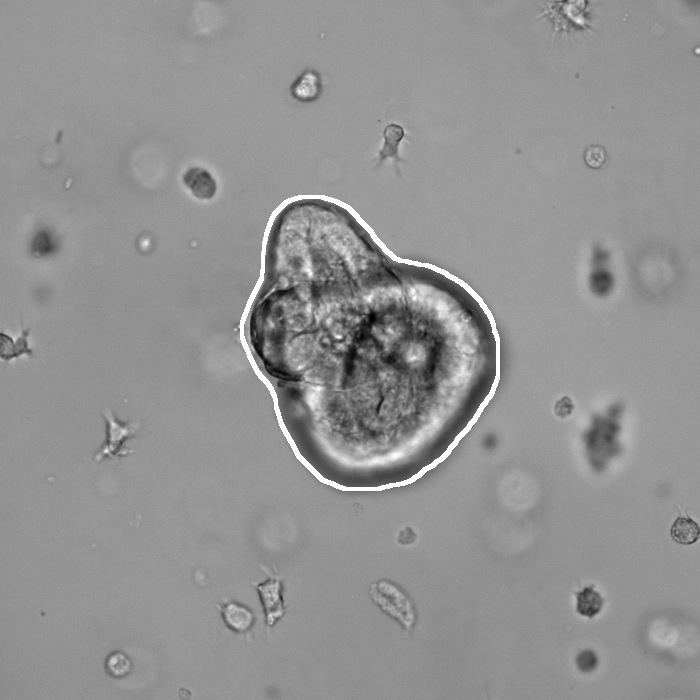
Resources
- The pre-trained model and source codes of our organoid analyzer [111 MB]
- The pre-trained model and source codes of our organoid texture simulator [1.0 GB]
- Real image data with manually created reference annotations [4.4 GB]
- Computer-generated image data with inherent reference annotations [2.5 GB]
Conditions of Use
The resources provided are free of charge for noncommercial and research purposes. Their use for any other purpose is generally possible, but solely with the explicit permission of the authors. In case of any questions, please do not hesitate to contact us at xmaska@fi.muni.cz.
In addition, you are expected to include adequate references whenever you present or publish results that are based on the resources provided.
References
- Lucia Hradecká, David Wiesner, Jakub Sumbal, Zuzana Sumbalova Koledova, Martin Maška. Segmentation and Tracking of Mammary Epithelial Organoids in Brightfield Microscopy. IEEE Transactions on Medical Imaging, 42(1):281-290, 2023.
Acknowledgment
This work was supported by the the Grant Agency of Masaryk University under the grant number MUNI/G/1446/2018 and by the Czech Science Foundation under the grant number GA21-20374S.