MRI Brain Screening Tool — Automated tumor-like pathology screening for clinical MRI

A friendly, research-grade software that automatically screens brain MRI exams (FLAIR / T2 / T1ce) and highlights tumor-like anomalies so radiologists can prioritise urgent cases.
Request evaluation
for non-commercial / research use only.
</div>
At a glance
- What it does: Automatically screens brain MRI exams and produces a short pre-diagnostic report (slice-level anomaly scores, highlighted slices, bounding boxes and optional segmentation masks).
- Clinical focus: Tumor-like pathologies (HGG, LGG, lymphoma, meningioma, metastases) in this first release.
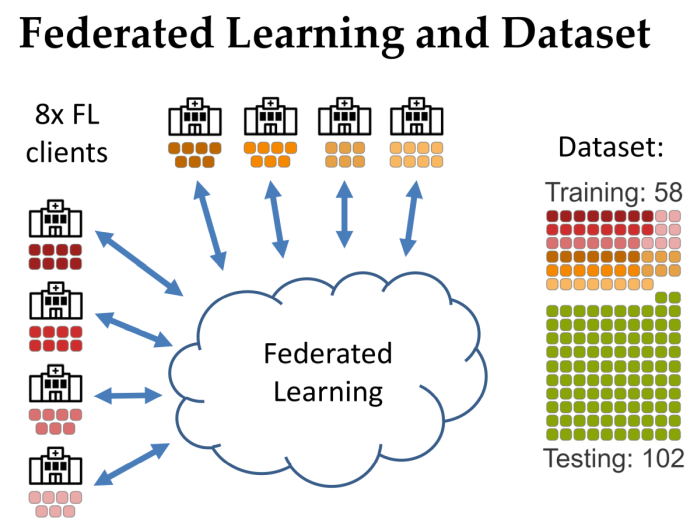
- Training approach: Federated Learning across multiple sites to enable collaborative model building without sharing patient data.
- Performance (validation on 102 exams): exam-averaged Dice ≈ 0.837, global Dice ≈ 0.884, sensitivity 0.98, precision 0.91.
- Availability: Non-commercial — available on request for evaluation/testing (contact: Roman Stoklasa).
Why researchers & clinicians use it
- Faster triage of urgent cases — flags exams likely to contain tumor-like pathology so radiologists can prioritize.
- High sensitivity for tumors — demonstrated high detection rate on the evaluation set, minimizing missed severe cases.
- Privacy-preserving collaboration — designed for Federated Learning so institutions contribute without exposing raw patient data.
- Lightweight, practical architecture — 2D slice U-Net with Inception-v3 encoder to reduce resource needs at client sites.
- Explainable output for radiologists — slice-level anomaly scores, a ranked set of most relevant slices and bounding boxes to support human review.
How it works — technical summary
- Input modalities: FLAIR, T2, T1ce (T1 can substitute if T1ce not available).
- Preprocessing pipeline: automatic skull-stripping → rigid/affine co-registration → intensity normalization (CDF/histogram matching to template) → resampling to common in-plane resolution (~0.72 mm/px).
- Model & inference: 2D axial slice processing with a U-Net (Inception-v3 backbone, pretrained encoder) that outputs binary lesion masks per slice; masks are fused to derive exam-level detections, anomaly scores and a ranked set of slices for review.
- Training: Federated averaging across multiple clients; heavy data augmentation with affine + elastic transforms; reported experiments used many federated rounds.
- Typical inference time: Example reported ~9 seconds per exam on a GTX 1080 + Intel i7-class CPU (performance depends on local hardware).
Evaluation & validation (concise)
- Dataset: A training dataset consisted of 58 examinations with a held-out validation/test set of 102 exams.
- Main metrics (reported on the held-out set):
- Exam-averaged Dice (EaD): 0.837.
- Global Dice (G.Dice): 0.884.
- Sensitivity (Recall): 0.98.
- Precision: 0.91.
Note: Comparison experiments showed federated training improved over very small local models and delivered performance comparable to strong baselines on the available dataset.
Technical specs (table)
| Item | Specification / notes | |—|—| | Supported input | FLAIR, T2, T1ce (T1 acceptable substitute) | | Model architecture | 2D U-Net with Inception-v3 encoder (pretrained weights) | | Preprocessing | Skull-strip, co-registration, intensity CDF-matching, resample to ~0.72 mm/px | | Training paradigm | Federated Learning (parameter averaging) across multiple clients; heavy augmentation | | Typical inference time | ≈ 9 s/exam on GTX 1080 + i7-class CPU (example) | | Validated pathologies | HGG, LGG, lymphoma, meningioma, metastases (tumor-like lesions) | | Validation set | 102 exams held-out (total dataset ≈160 exams) | | Output | Pre-diagnostic report: slice-level scores, ranked slices, bounding boxes, optional segmentation masks | | Intended users | Radiology departments, research groups, multi-site federated ML projects |
Deployment & requirements
- Server / hardware: Flexible — inference is lightweight. Full GPU recommended for fastest throughput.
- Integration: The tool can run in batch mode (overnight processing) or inline after acquisition; reports can be delivered via standard channels (file drop, email, PACS connector).
- Privacy & collaboration: Built to support federated orchestration so institutions keep raw data on-premise and only share model updates.
Use cases
- Triage: Flag likely severe cases so they are read earlier.
- Quality assurance: Double-check routine reads to catch missed anomalies.
- Research / federated projects: Join or form federations to collaboratively train improved models while keeping local data private.
Pricing & availability
- License / price: Non-commercial research release. The tool is the result of academic research and is available on request for evaluation/testing. Contact the corresponding author for evaluation access.
Sample outputs (what the radiologist sees)
- Ranked list of “most interesting” axial slices with anomaly scores.
- Axial / sagittal anomaly score plots to indicate location (left / right) of abnormality.
- Bounding boxes shown on slices; optional precise segmentation overlay available if requested.

Limitations & caveats
- Current pathology scope: Focused on tumor-like lesions — detection of other pathologies (WMH, MS, stroke) is not the primary target for this release.
- Dataset size & generalisability: The current model was trained on a relatively small training set; local retraining or federated participation is recommended for best local performance.
- Regulatory status: Research release — not cleared for diagnostic decision-making. Clinical deployment requires local validation and regulatory review.
FAQ (short)
Q1 — Can the tool replace a radiologist?
No. It produces pre-diagnostic reports to help prioritise and flag exams; final diagnosis remains with a qualified radiologist.
Q2 — What MRI sequences are required?
FLAIR, T2 and T1ce are the designed inputs. T1 (non-contrast) can be used if T1ce is missing.
Q3 — How accurate is it on tumors?
On the reported evaluation it achieved high sensitivity (≈98%) with high precision (≈91%) and global Dice >0.88 on the held-out set.
Q4 — Is patient data shared during federated training?
No. Only model updates are shared; raw images remain on the local site.
Q5 — Can we run it offline / behind hospital firewall?
Yes — the model and preprocessing can be run locally; federated participation requires networked aggregation only for training updates.
Q6 — How do we request an evaluation copy?
Contact the presenting author: Roman Stoklasa — stoklasa@fi.muni.cz. Include your institution, intended use (evaluation / research), and technical contact.
References
If you use this tool, please cite the following paper:
- Stoklasa R, Stathopoulos I, Karavasilis E, Efstathopoulos E, Dostál M, Keřkovský M, Kozubek M a Serio L. Brain MRI Screening Tool with Federated Learning. Online. In 2024 IEEE International Symposium on Biomedical Imaging. Athens, Greece: IEEE, 2024, s. 1-5. ISBN 979-8-3503-1333-8. Available from: https://dx.doi.org/10.1109/ISBI56570.2024.10635396.
Acknowledgement
This project was partially funded by the CERN budget for Knowledge Transfer for the benefit of Medical Applications, by the Ministry of Health of the Czech Republic (Grant No. NU21-08-00359) and the Ministry of Education, Youth and Sports of the Czech Republic (Project No. LM2023050).
Credits
- Roman Stoklasa, Ph.D. (main author and developer, federated learning, data pipeline, software architecture and its )
- Ioannis Stathopoulos (research and experimentation with anomaly detection and classification in MRI brain images)