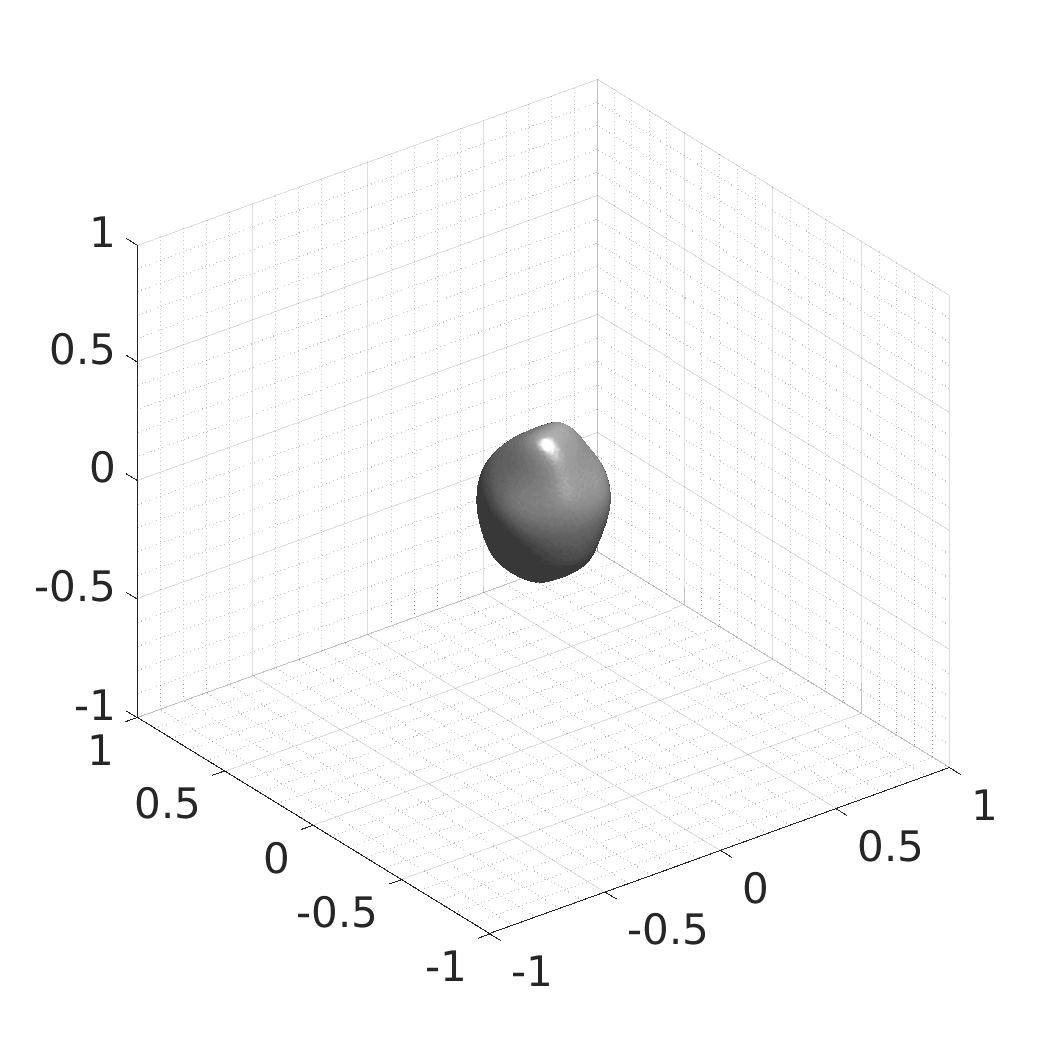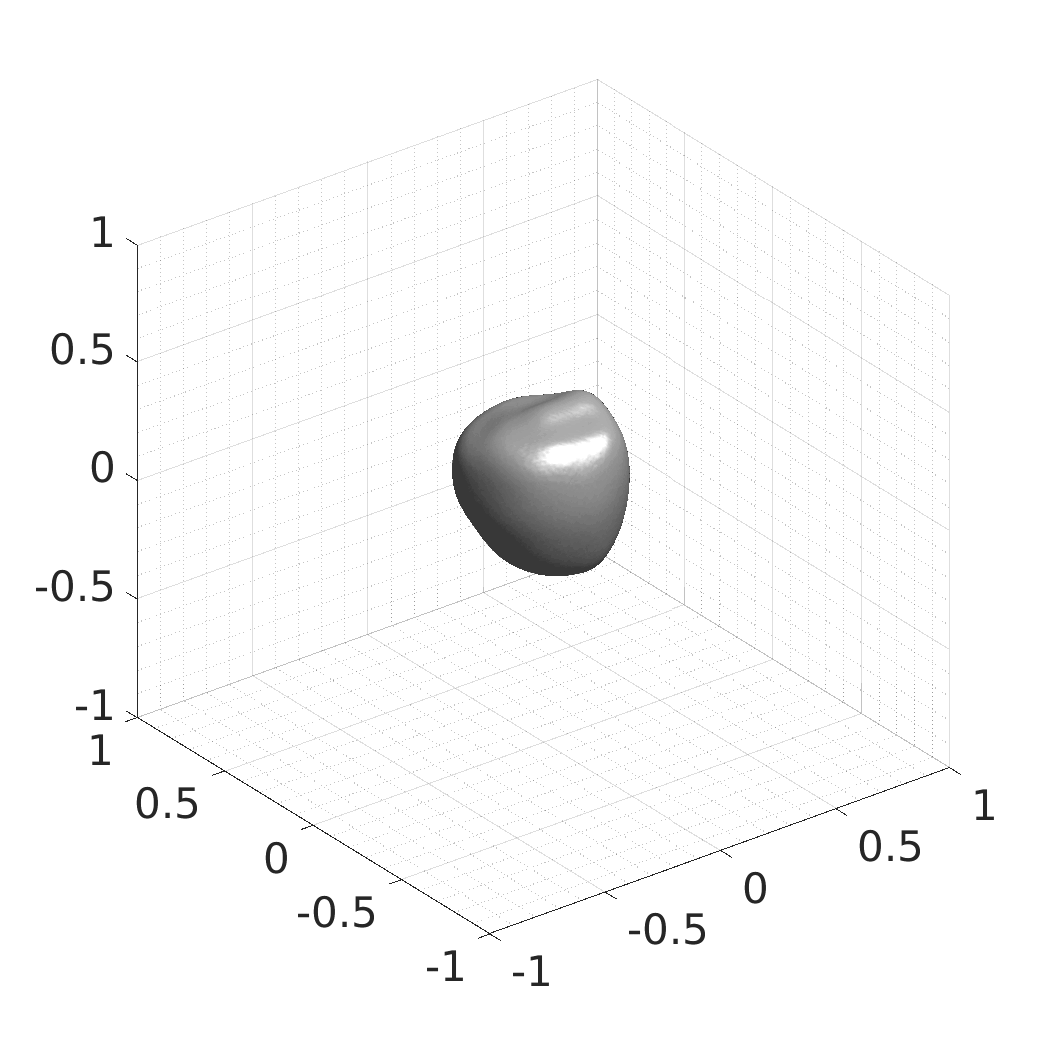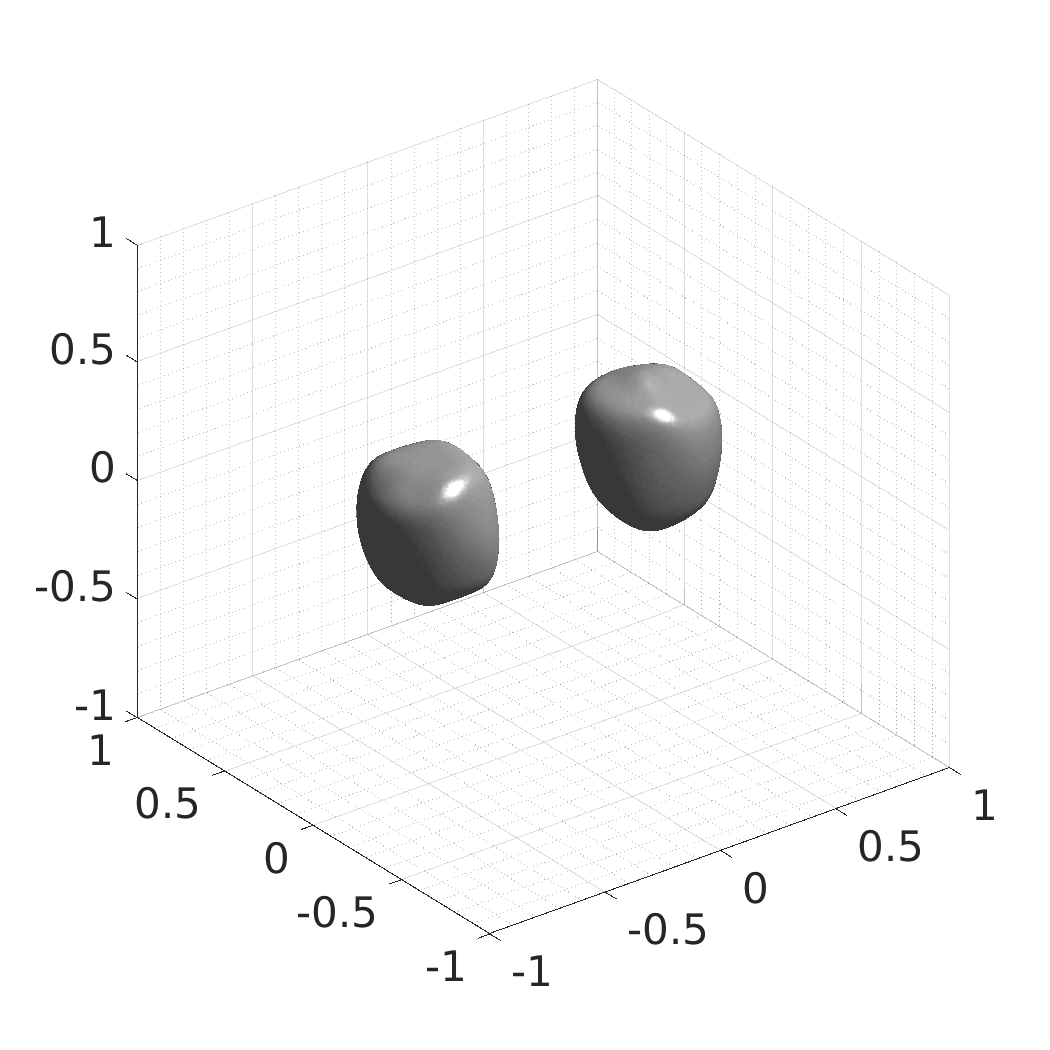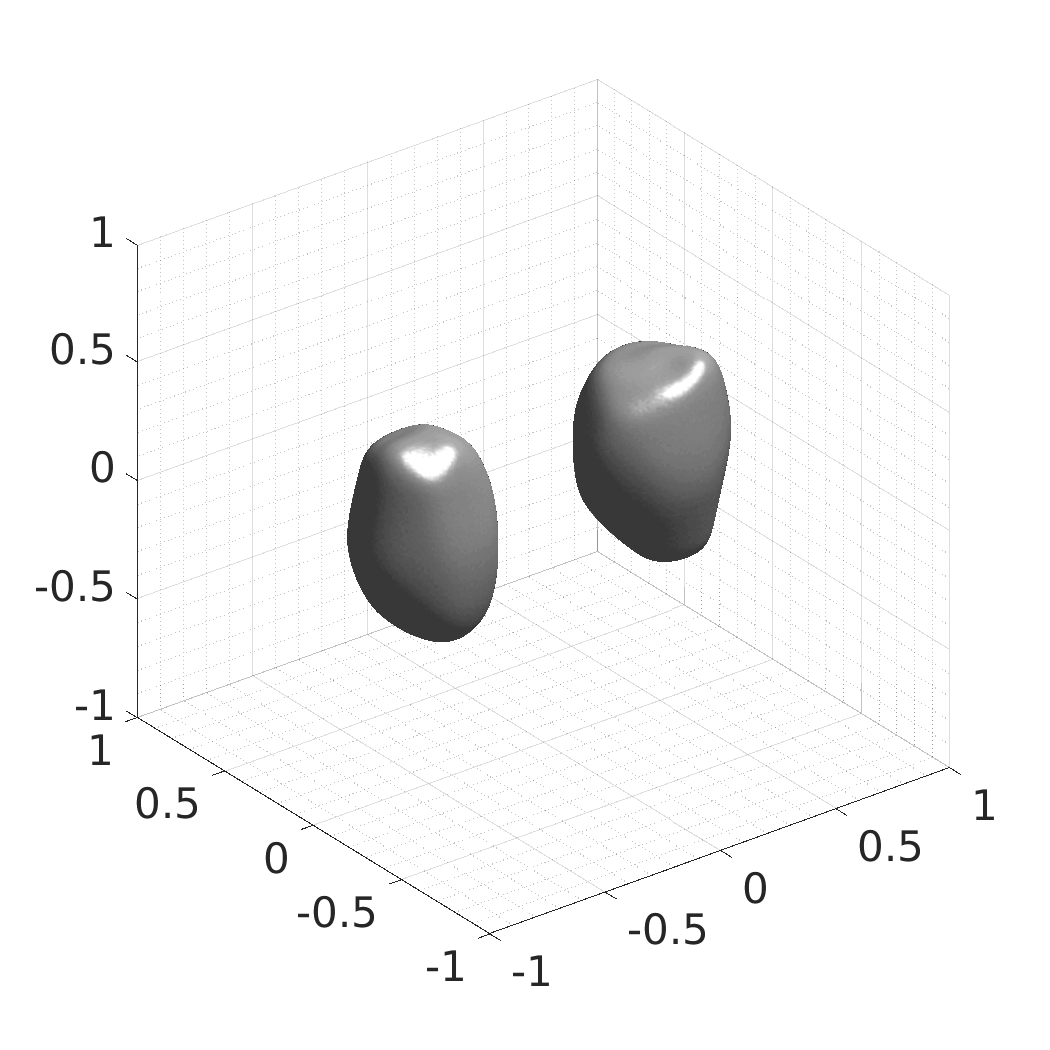
Generative modeling of living cells with implicit neural representations
David Wiesner, Julian Suk, Sven Dummer, Tereza Nečasová, Vladimír Ulman, David Svoboda, and Jelmer M. Wolterink
Journal paper | Conference paper | Conference slides | Conference poster | Source code (GitHub)
This is the official web page of MedIA 2024 paper “Generative modeling of living cells with SO(3)-equivariant implicit neural representations” and MICCAI 2022 paper “Implicit Neural Representations for Generative Modeling of Living Cell Shapes”. Here, we make available the complete source code, pre-trained models, and data sets presented in the paper.
Introduction
Contemporary methods for cell shape synthesis in biomedical imaging often sample from a pre-determined database or utilize simple geometric objects like ellipsoids. These methods considerably narrow down the variability in the synthetic images. In addition, they are usually difficult to adapt to different cell types, which limits their usability. The methods utilizing voxel-based representations are further limited by the computational and memory demands that prohibit generating shapes at high spatial and temporal resolutions.
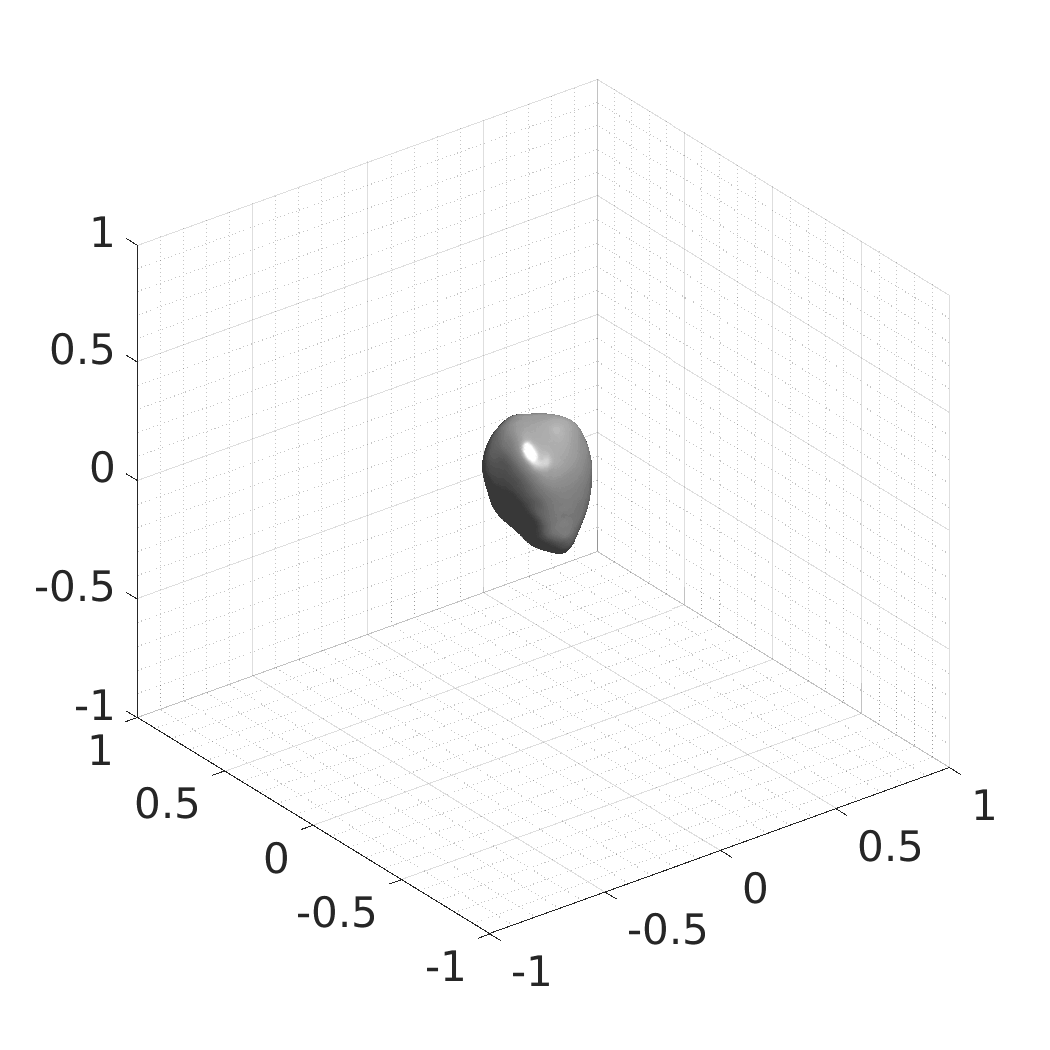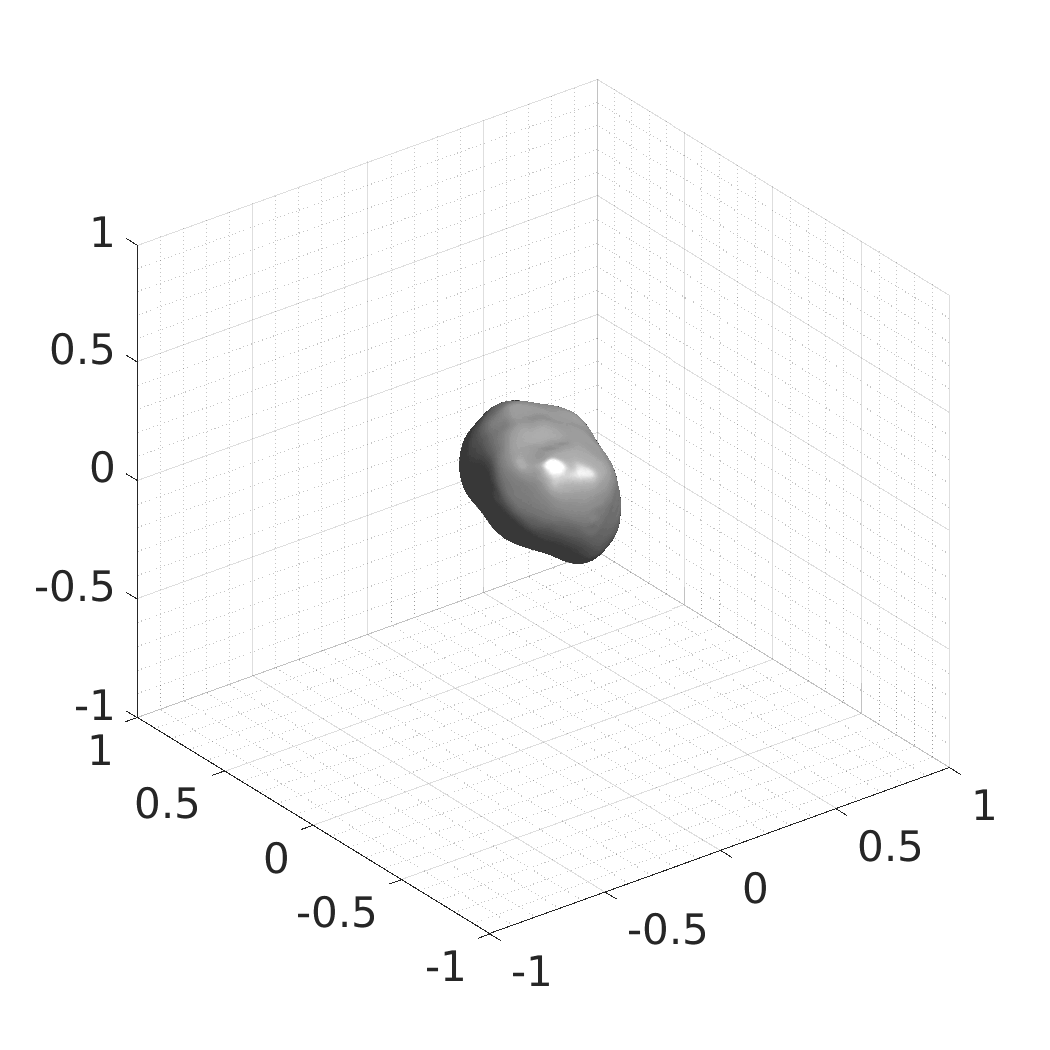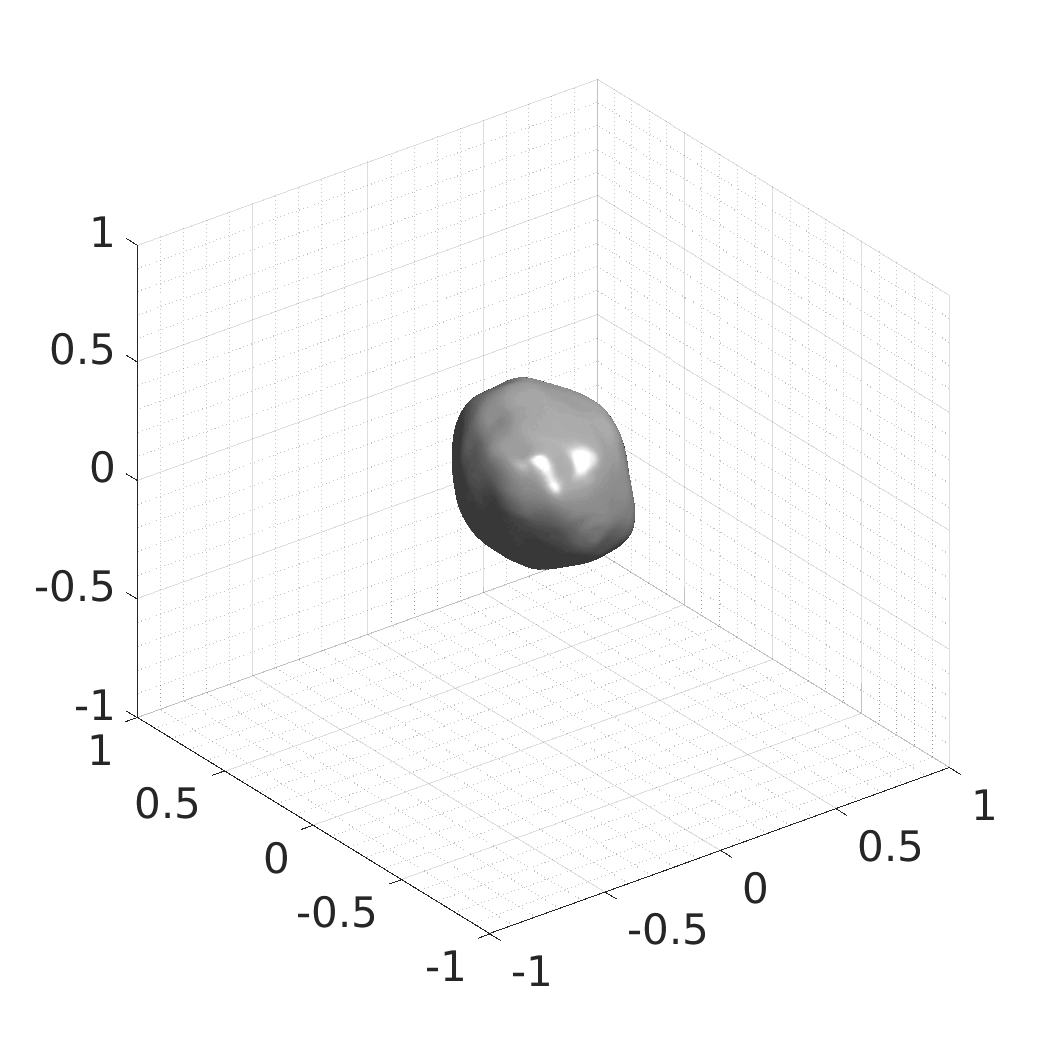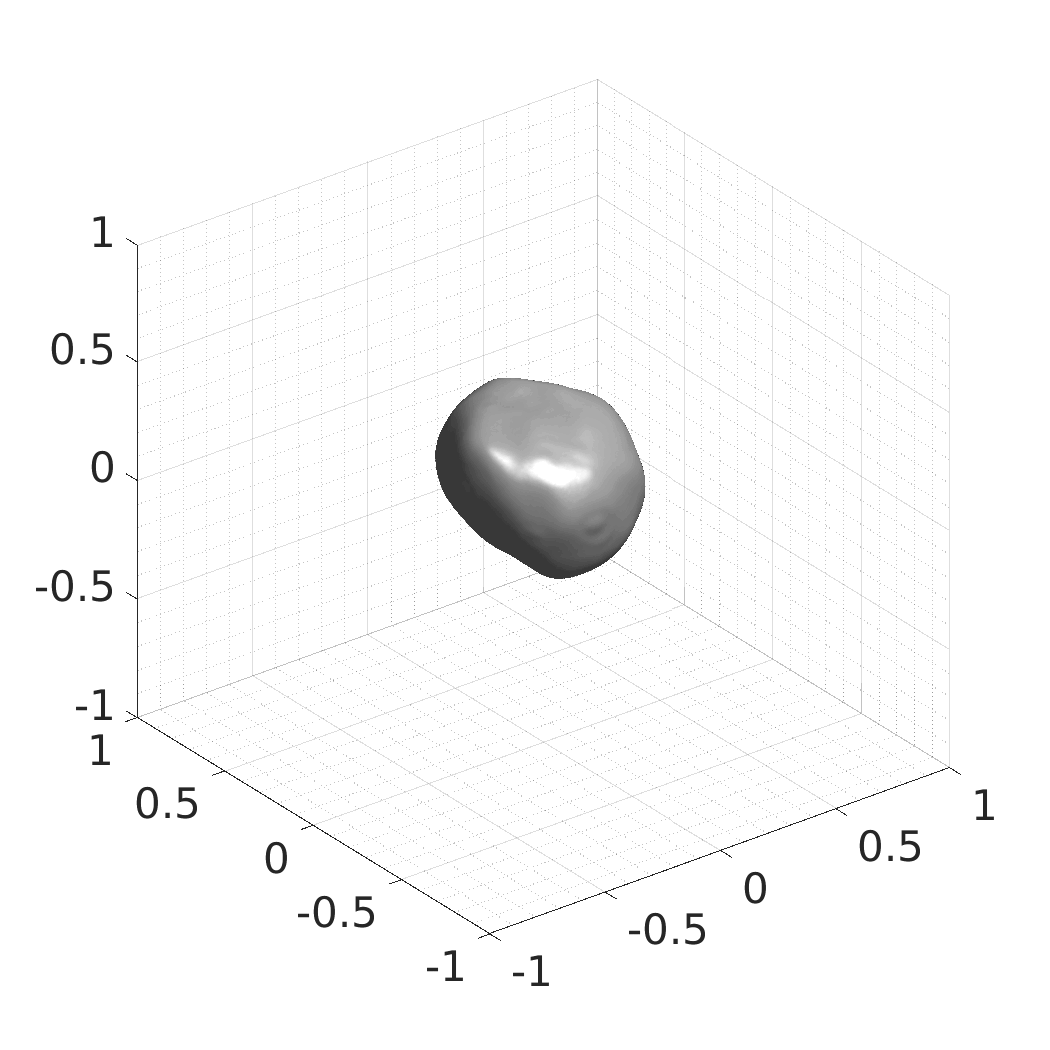
In this work, we optimize a neural network as an implicit neural representation of arbitrary time-evolving living cell shapes. We define these time-evolving cell shapes by 3D+time signed distance functions (SDF), rather than voxel volumes directly. The SDF representation allows for non-uniform sampling, which facilitates representing a complex 3D scene with a much lower number of data points than traditional voxel-based representations. Instead of traditional ReLU activations, we utilize periodic functions, which exhibit faster convergence and allow for fitting high frequency shapes with sharp edges. We demonstrate our approach on three distinct cell lines: the Platynereis dumerilii embryo cells that exhibit rapid growth and shape deformations, the C. elegans embryo cells that grow and divide, and the A549 human lung carcinoma cells with growing and branching filopodial protrusions.
## Implementation of the Method
The following guide is applicable for Linux-based systems. The versions of the libraries and command line parameters may slightly differ for Windows or macOS systems.
Requirements and Dependencies
The implementation was tested on AMD EPYC 7713 64-Core Processor, 512 GB RAM, NVIDIA A100 80 GB GPU, and Ubuntu 20.04 LTS with the following versions of software:
- NEURAL NETWORK
- Python 3.9.16
- PyTorch 2.0.1
- PyTorch3D 0.7.4
- NumPy 1.25.1
- SciPy 1.11.1
- tqdm 4.65.0
- h5py 3.9.0
- Spyder 5.4.3 (optional)
- DATA PROCESSING AND VISUALIZATION
- Matlab R2022a
- DIPimage 2.9 (optional)
Downloads
- Source code, pre-trained models, and examples (1.2 GB)
- Full training data set - Platynereis dumerilii [HDF5] (8.2 GB)
- SDFs of 33 time-evolving cell shapes at 30 time points with a grid of 256×256×256.
- Full training data set - C. elegans [HDF5] (9.1 GB)
- SDFs of 33 time-evolving cell shapes at 30 time points with a grid of 256×256×256.
- Full training data set - A549 filopodial [HDF5] (3.5 GB)
- SDFs of 33 time-evolving cell shapes at 30 time points with a grid of 256×256×256.
- Full training data set - Rotated Platynereis dumerilii [HDF5] (6.7 GB)
- SDFs of 100 time-evolving cell shapes at 8 time points with a grid of 256×256×256. There are 25 Platynereis dumerilii shapes, each randomly rotated four times over X, Y, and Z axes.
- Full training data set - All cell types [HDF5] (5.5 GB)
- SDFs of 99 time-evolving cell shapes at 8 time points with a grid of 256×256×256. There are 33 Platynereis dumerilii shapes, 33 C. elegans shapes, and 33 A549 filopodial shapes.
Quick Start Guide
To follow this guide, please download and extract the Source code, pre-Trained models, and examples (1.2 GB) and optionally the training data sets.
-
Installing the Conda Environment (Optional)
We prepared a pre-configured conda environment with all required libraries for the generative model. Conda is available here. After setting up Conda, you can install the required environment using the included./autodecoder/conda_env.ymlfile:
$> conda env create -f conda_env.yml -
Shape Reconstruction
To reconstruct the learned shape SDFs using the pre-trained models, execute the script./autodecoder/test.pywith parameters specifying the desired model directory (whereplatstands for Platynereis dumerilii cells,celefor C. elegans cells, andfilofor filopodial cells):
$> python test.py -x experiments/<model> -t reconstruct
The resulting SDFs in MAT or HDF5 format will be saved in./autodecoder/experiments/<model>/OUT_reconstruct. You can use the Matlab script./autodecoder/experiments/<model>/quick_preview.mto get PNG bitmaps previewing the resulting SDFs. -
Inferring New Shapes
New SDFs are produced using randomly generated latent codes (in the case of C. elegans and Platynereis dumerilii), or by adding noise to the learned latent codes (in the case of A549 filopodial cells). To infer new SDFs using the pre-trained models, execute the script./autodecoder/test.pyand specify the appropriate model directory:
$> python test.py -x experiments/<model> -t generate
For A549 filopodial cells, use this command:
$> python test.py -x experiments/filo -t generate_filo
The resulting SDFs in MAT or HDF5 format will be saved in./autodecoder/experiments/<model>/OUT_randomgen. You can use the Matlab script./autodecoder/experiments/<model>/quick_preview.mto get PNG bitmaps previewing the resulting SDFs. -
Training the Network
To train the network, download one of the training data sets and extract it to./autodecoder/datafolder. The training SDFs are represented by 4D single precision floating point arrays in HDF5 format. The configuration files./autodecoder/experiments/<model>/specs.jsoncontain pre-defined training parameters. To train the model, execute the./autodecoder/train.pyscript and specify the desired model directory:
$> python train.py -x experiments/<model>
Please note that the training parameters in the providedspecs.jsonfiles are configured for GPUs with 80 GB of memory. To reduce the memory consumption, you can edit the configuration to reduce the number of time points in a training batchFramesPerBatchor the number of SDF points sampled per time pointTrainSampleFraction. After training, you can test the resulting model using:
$> python test.py -x experiments/<model> -t reconstruct -e <epoch> -
Preparing Your Own Training Data Sets (Matlab + CytoPacq)
You can use 3D voxel volumes of shapes to prepare new training SDFs. An example Matlab script./matlab/prepare_training_data/voxvol_to_sdf.muses synthetic cells generated using the CytoPacq web-interface, available at https://cbia.fi.muni.cz/simulator, to prepare training data. Basic preprocessing steps, such as shape centering and checking the number of connected components, are implemented. Supported output formats for the SDFs are MAT and HDF5. We recommend using HDF5 for larger data sets due to stronger compression and the support of data larger than 2 GB. This script expects synthetic data sets generated using CytoPacq but can be modified to suit your specific needs. Three time-evolving shapes with 30 time points generated using CytoPacq are included as an example. -
Spatial and Temporal Interpolation
The trained neural network constitutes a continuous implicit representation of the SDFs and thus is able to produce outputs in arbitrary spatial and temporal resolution. The spatial interpolation can be used to increase spatial resolution of the shapes, whereas the temporal interpolation can be used to increase the number of time points. The interpolation does not require re-training the network and can be configured by adjusting the respective parameters inspecs.json. For spatial interpolation, set the parameterReconstructionDims, and for temporal interpolation, set the parameterReconstructionFramesPerSequence. The interpolation is applicable for reconstruction or random generation of new shapes.
## Produced Datasets We used the optimized neural network and randomly sampled latent codes to produce new spatio-temporal SDFs of evolving shapes. A separate model was optimized for each cell line. The produced SDFs were converted to binary voxel volumes representing the cell shapes. These shapes were subsequently textured using a conditional generative model to produce synthetic datasets suitable for benchmarking of image analysis algorithms.
Randomly Generated Synthetic Cell Shapes
Each of the following archives contains 33 synthetic 3D time-lapse sequences of the respective cell line that were generated using the proposed method, where each sequence captures an evolving cell shape at 30 time points. The cell shape at each time point is represented by a binary volume of resolution 256×256×256 voxels that is saved as lossless TIFF.
- Filopodial Cell Shapes [3D TIFF] (38 MB)
Synthetic Segmentation Benchmark
The benchmark datasets were produced using the synthetic cell shape sequences discussed in the previous section. We computed maximum intensity projections of the 3D voxel volume at each respective time point to obtain binary masks. After that, we used a conditional generative model pix2pixHD, which was trained on real images acquired using a microscope, to produce plausible-looking grayscale textures for these masks. The obtained masks serve as a ground truth for segmentation of the synthetic microscopy images and thus are suitable for benchmarking of image analysis algorithms. Each archive contains 33 2D time-lapse sequences, with each time point represented by a pair of synthetic texture and the corresponding binary mask, having resolution of 256×256 pixels and saved as lossless TIFF.
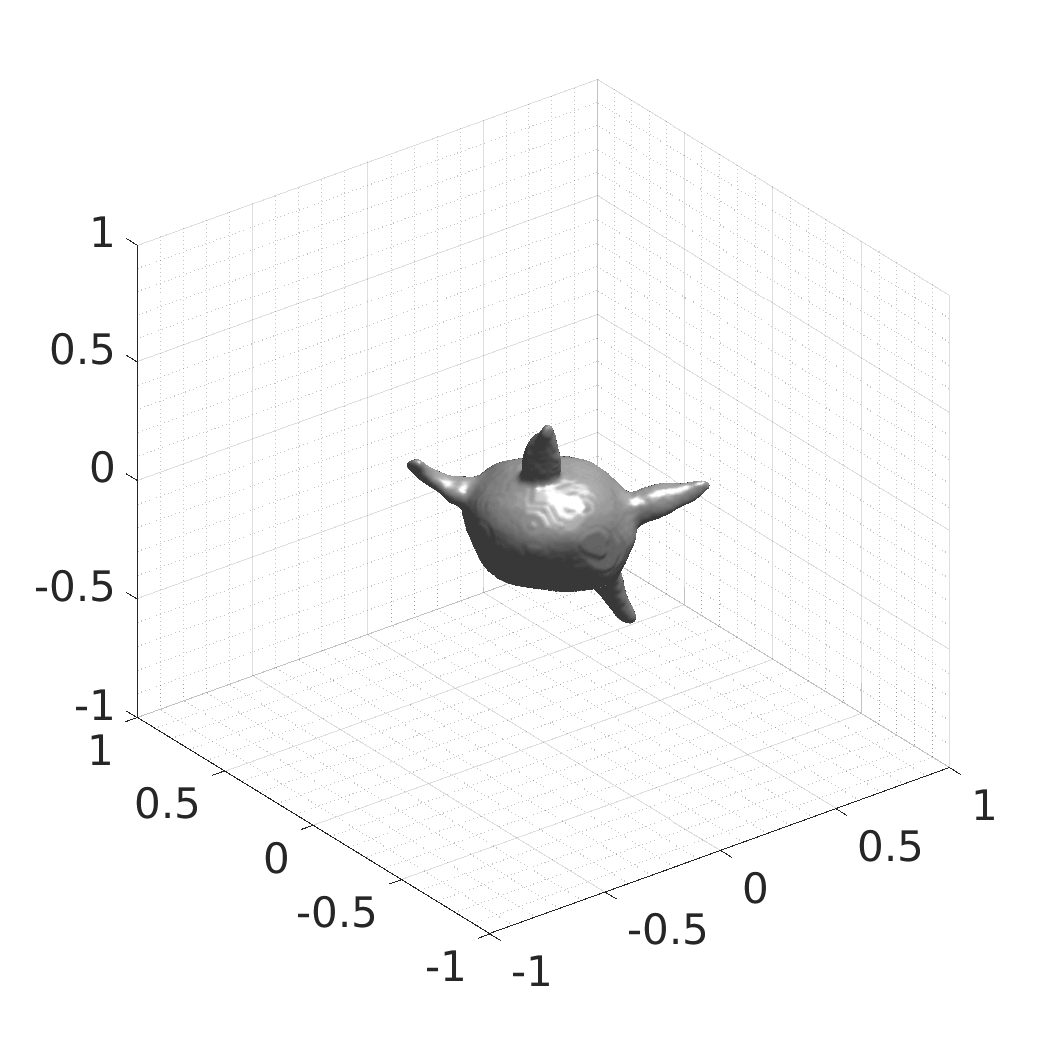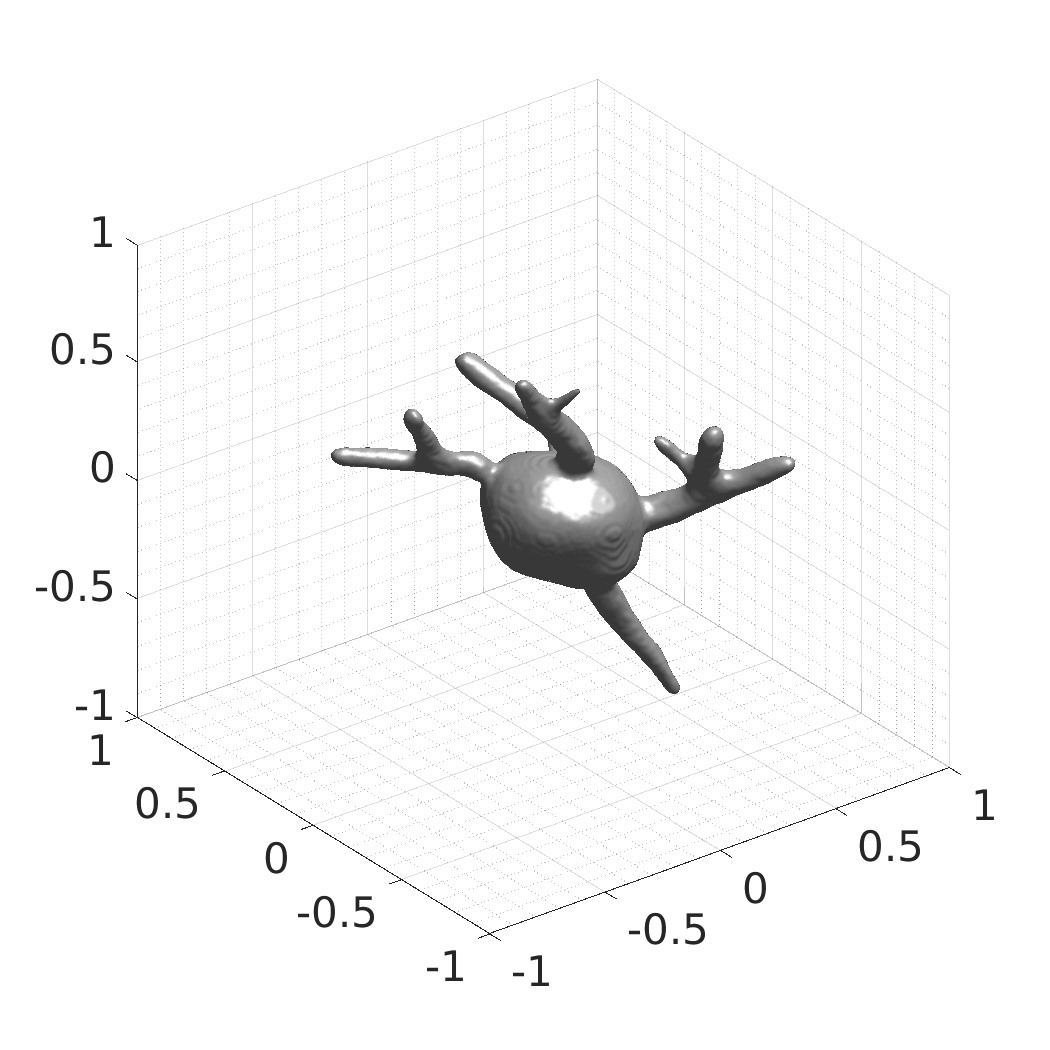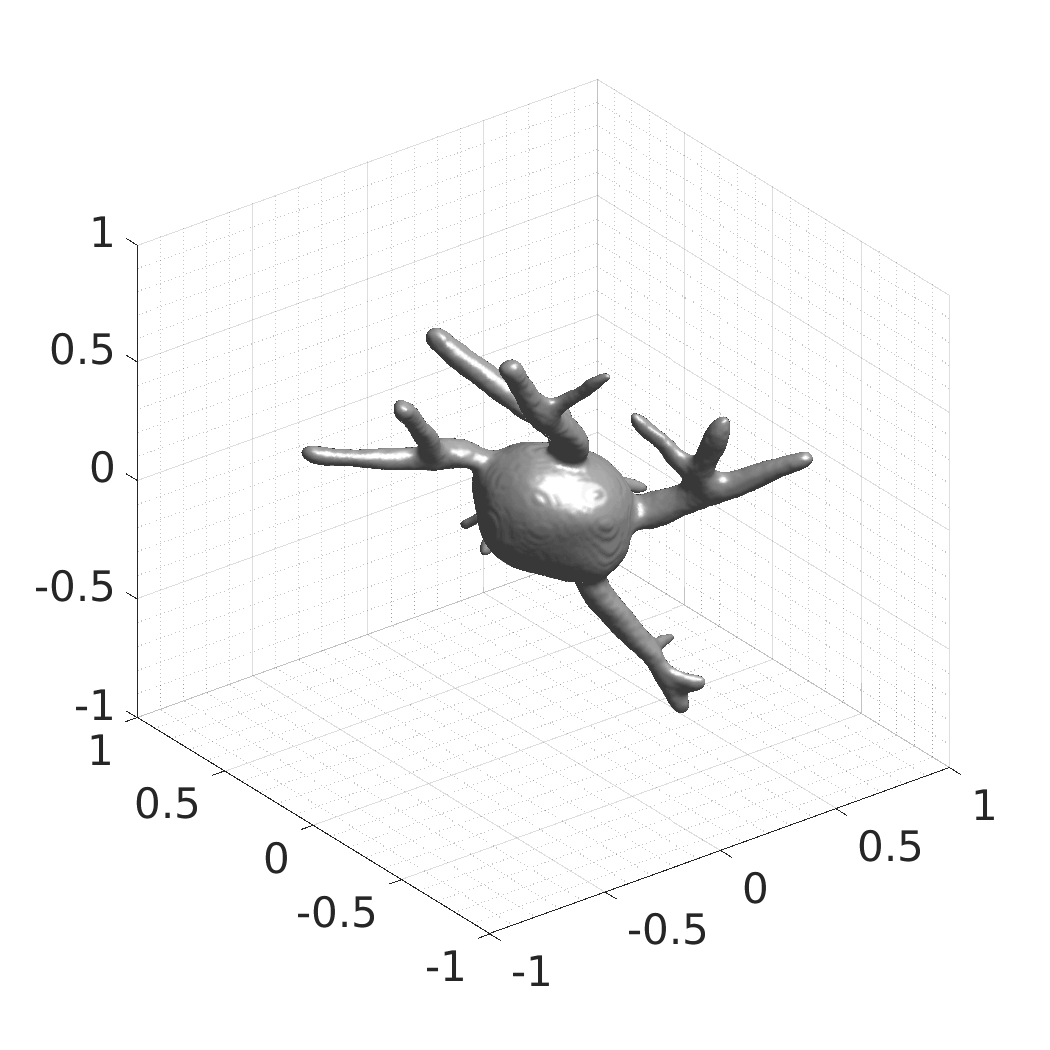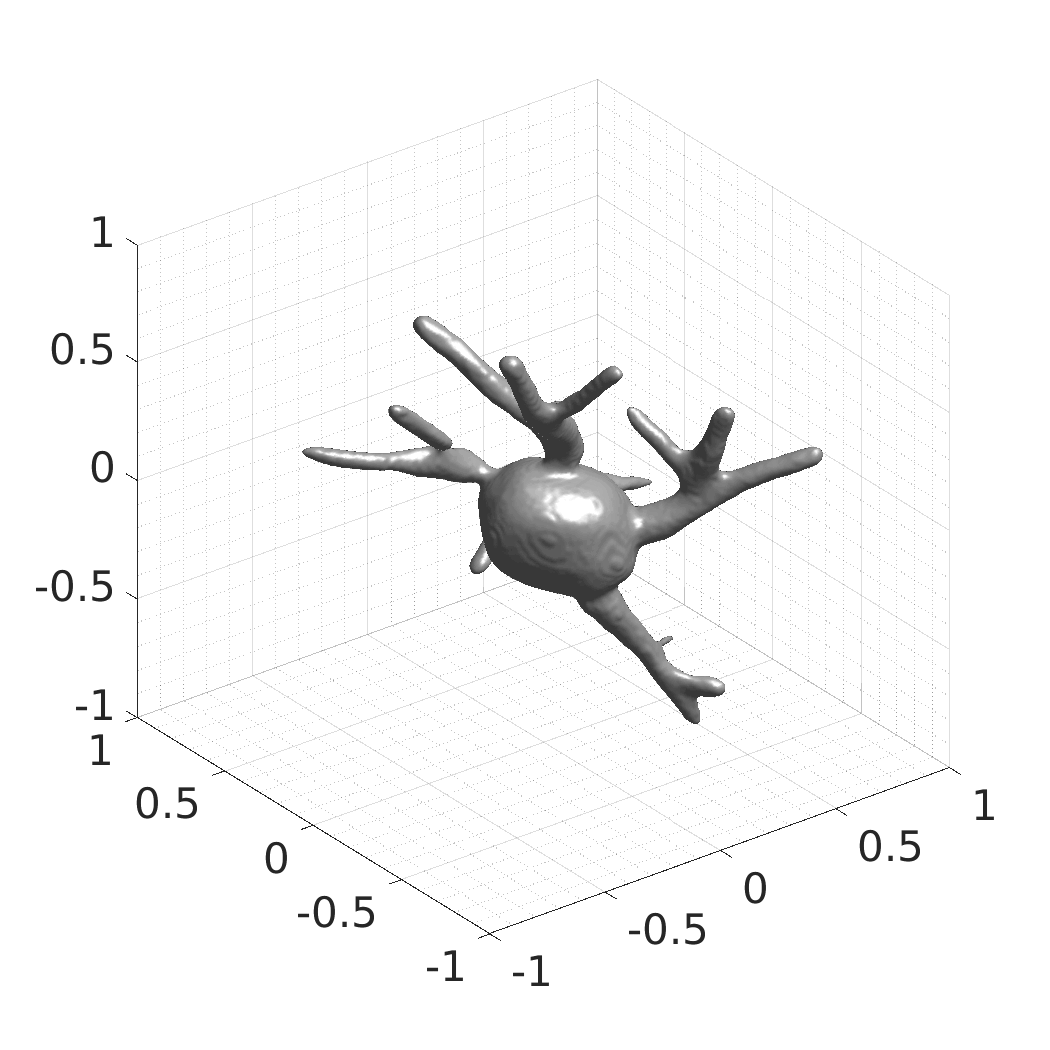
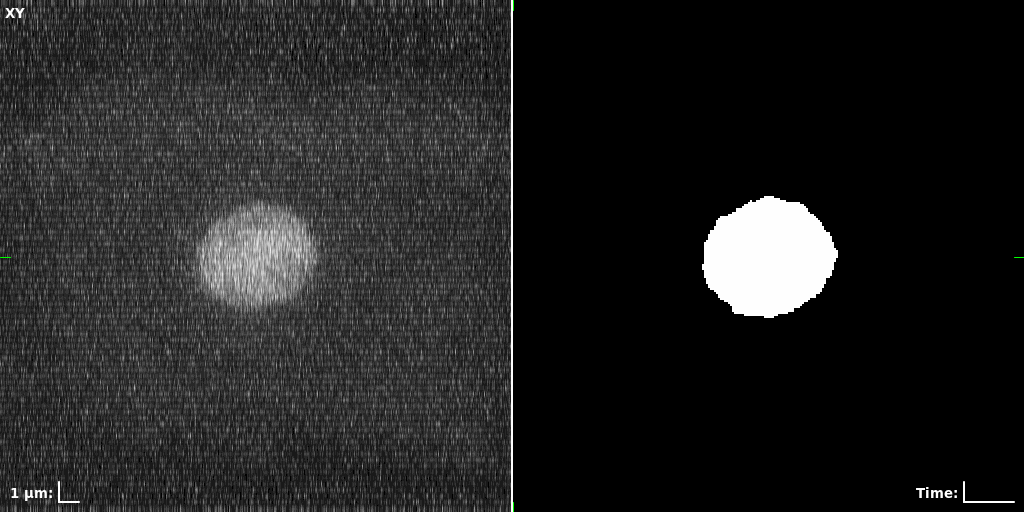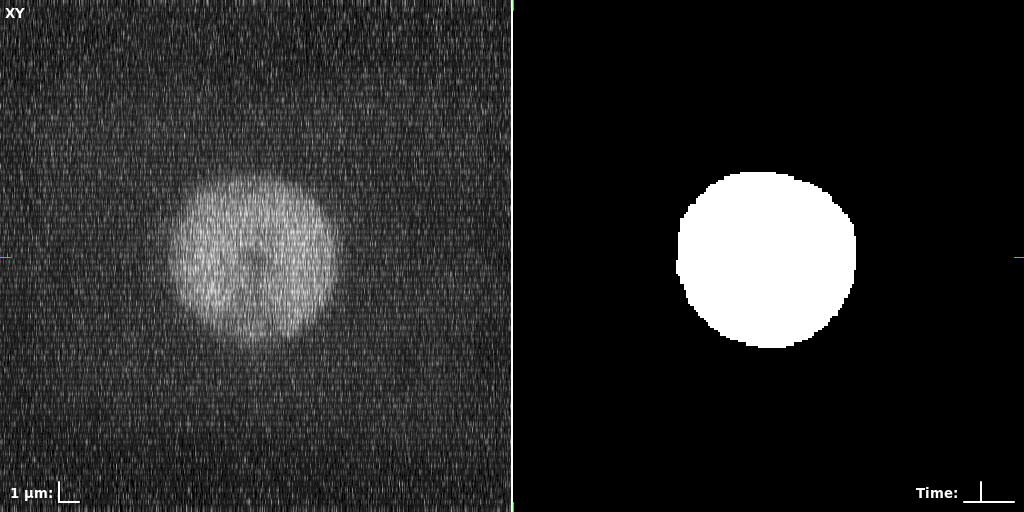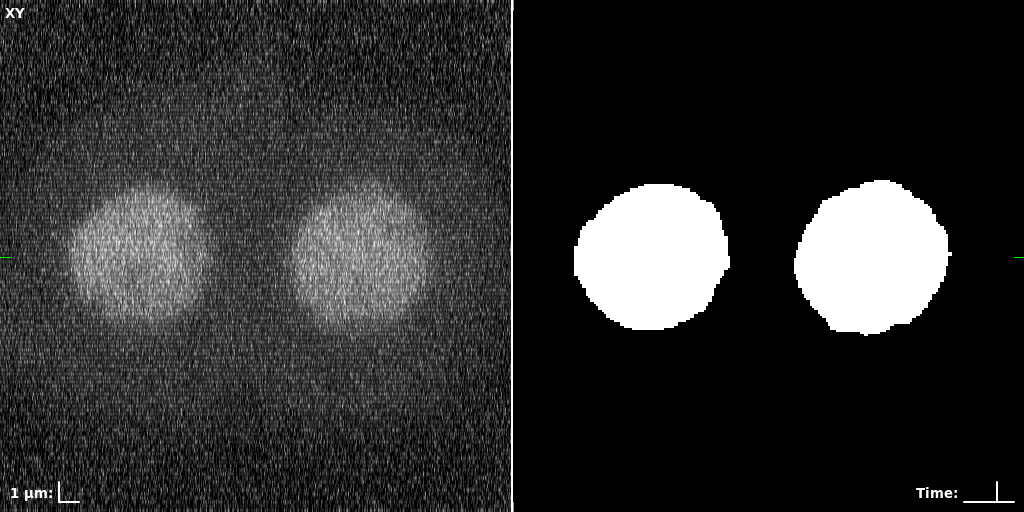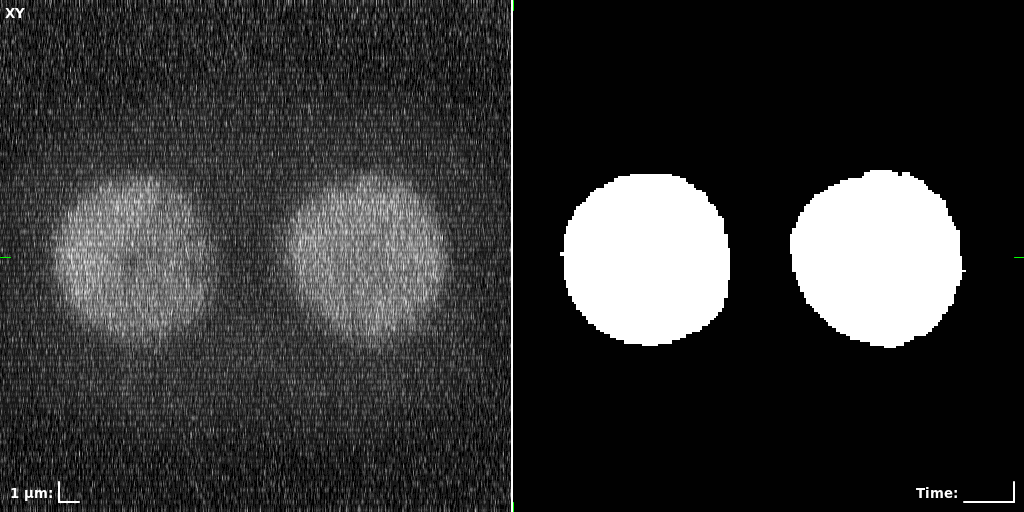
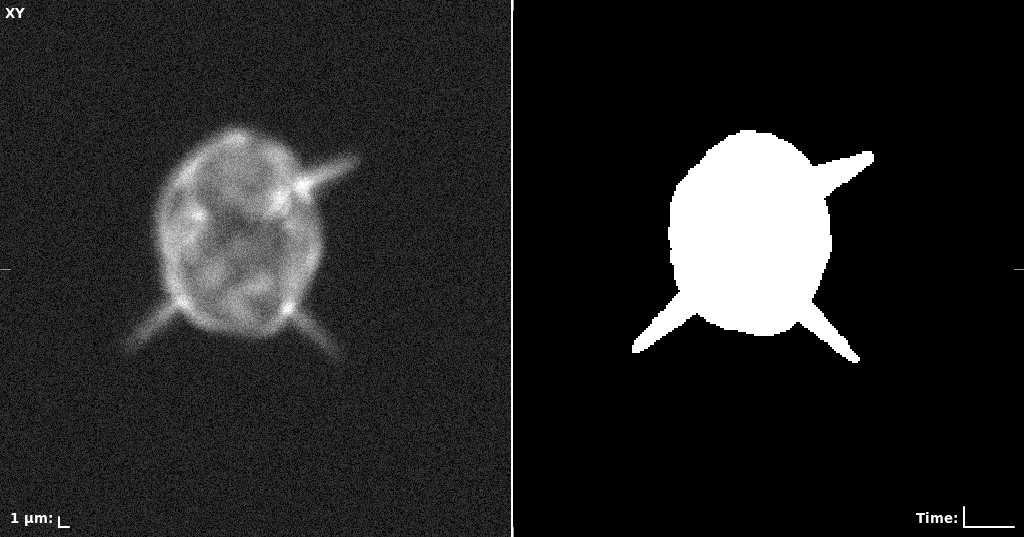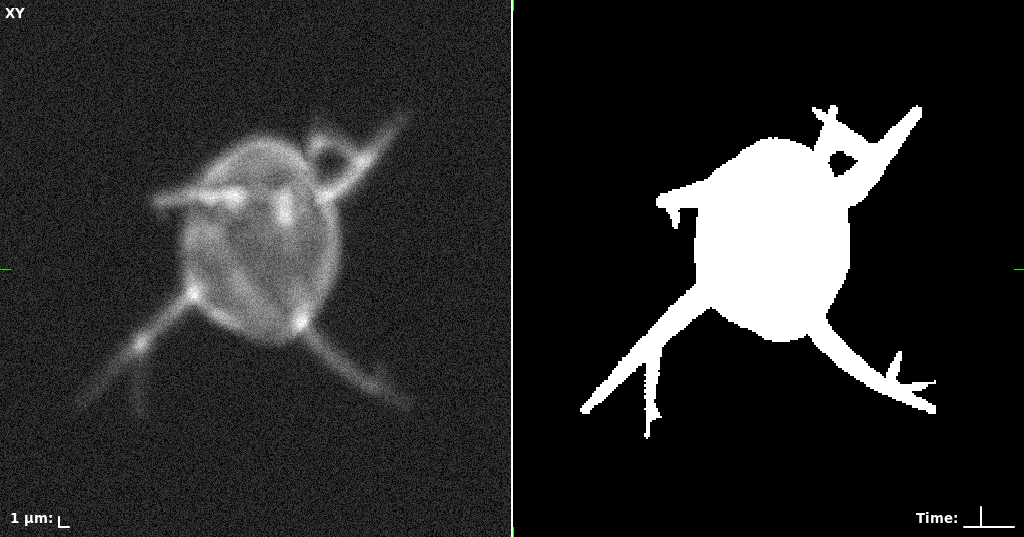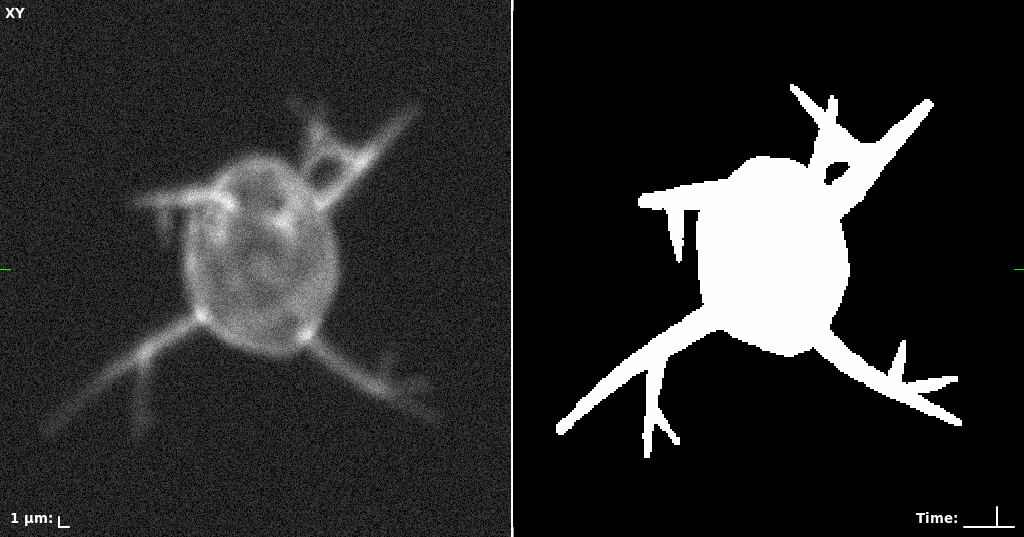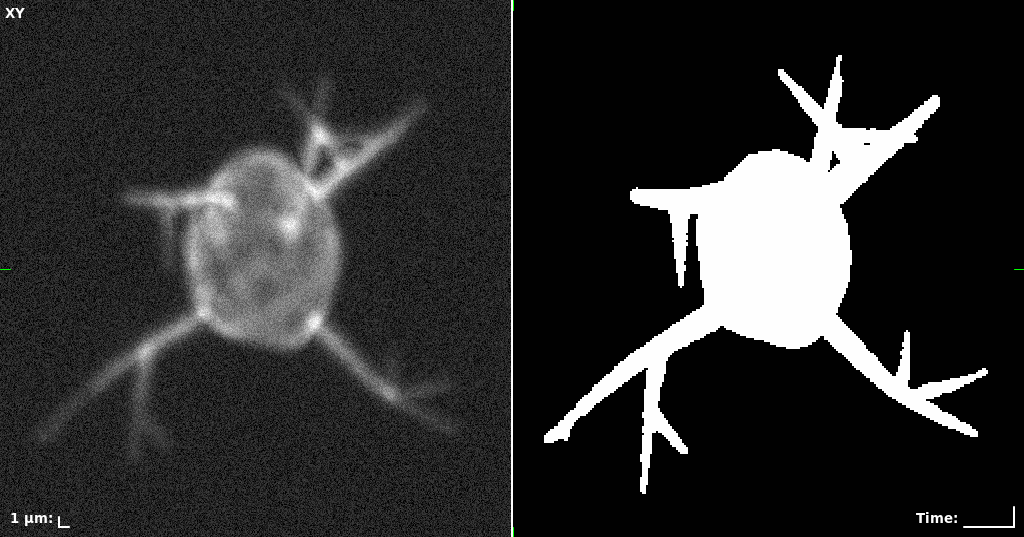
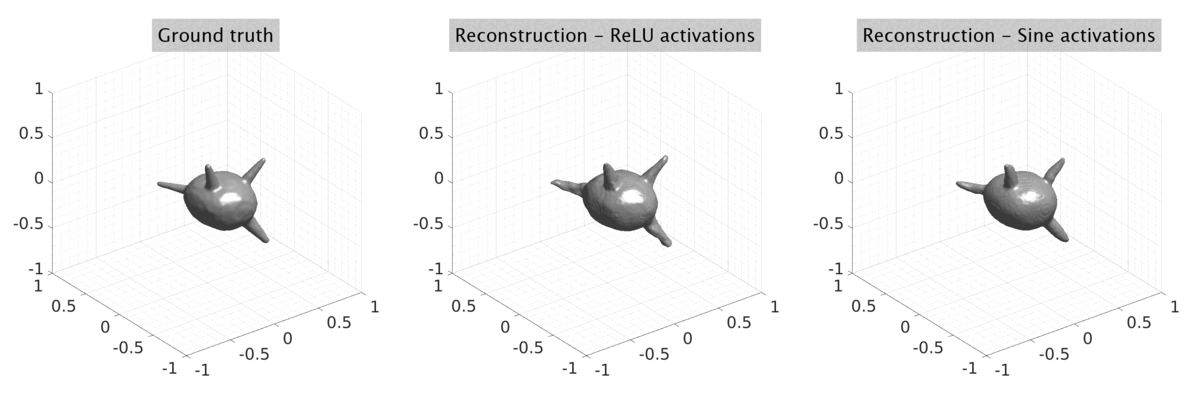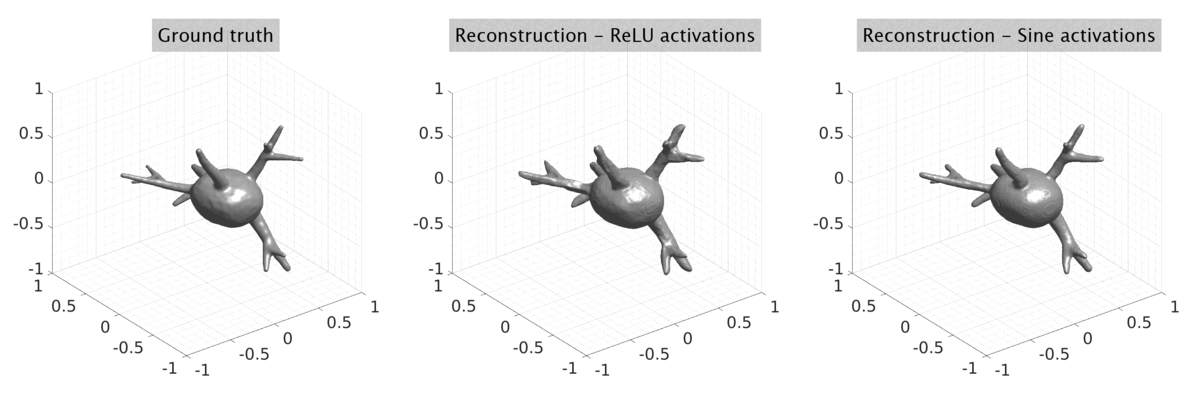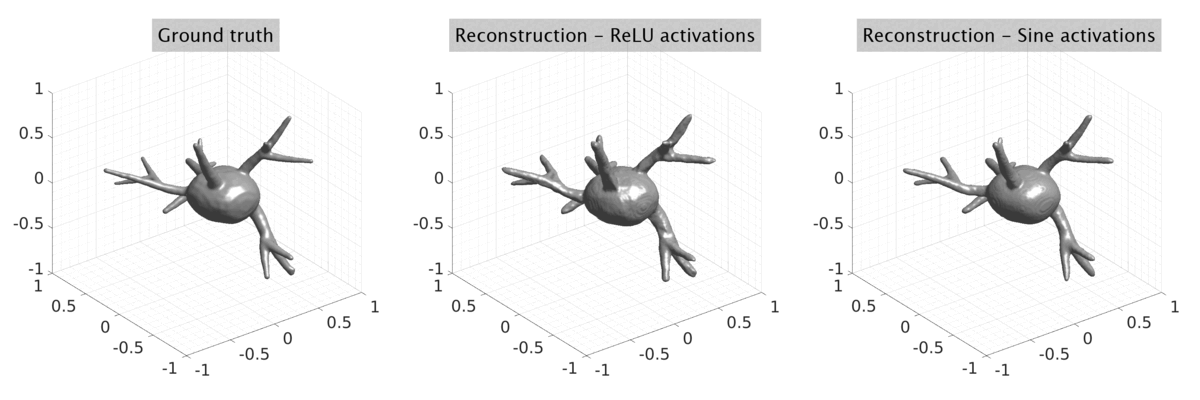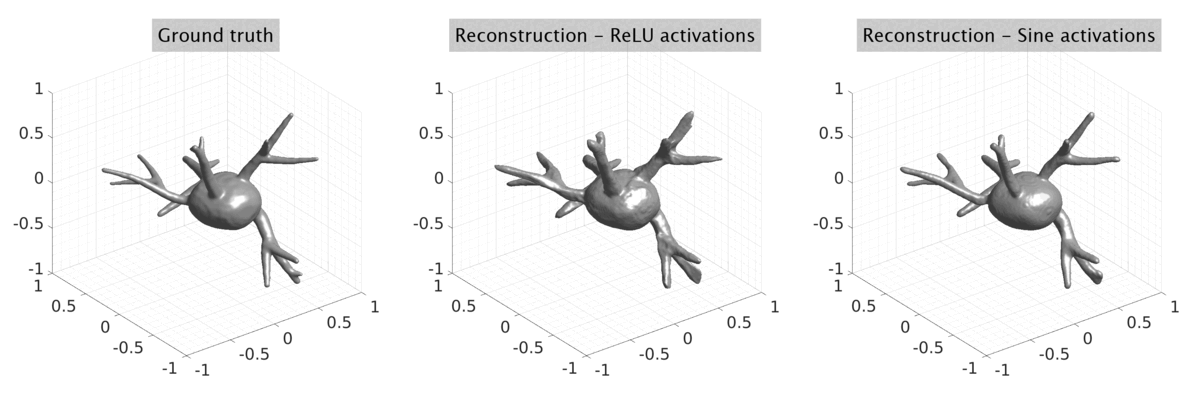
## Visual Comparison Between Periodic and ReLU Activation Functions
Here, we present a visual comparison of a single filopodial cell reconstructed with the models using periodic activation functions (sine) and ReLU activation functions. The models were trained to fit the ground truth sequence and optimize a latent code representing this time-evolving cell shape. After that, the latent code was used for reconstruction of the cell shape. Notice that the time-evolving shape reconstructed using the ReLU activations exhibits perceivable noise on the cell surface that is not present in the ground truth sequence, and that the respective filopodial protrusions are thicker and less detailed.
## Citation
If you find our work useful in your research, please cite:
-
Journal paper
Wiesner D, Suk J, Dummer S, Nečasová T, Ulman V, Svoboda D and Wolterink J.M. Generative modeling of living cells with SO(3)-equivariant implicit neural representations. Medical Image Analysis. 2024, vol. 91, p. 102991. ISSN 1361-8415. doi:10.1016/j.media.2023.102991.BibTeX:@article{wiesner2024media,
title={Generative modeling of living cells with {SO}(3)-equivariant implicit neural representations},
author={Wiesner, David and Suk, Julian and Dummer, Sven and Ne{\v{c}}asov{\'a}, Tereza
and Ulman, Vladim{\'\i}r and Svoboda, David and Wolterink, Jelmer M.},
journal={Medical Image Analysis},
volume={91},
pages={102991},
year={2024},
issn={1361-8415},
doi={https://doi.org/10.1016/j.media.2023.102991},
url={https://www.sciencedirect.com/science/article/pii/S1361841523002517}
} -
Conference paper
Wiesner D, Suk J, Dummer S, Svoboda D and Wolterink J.M. Implicit Neural Representations for Generative Modeling of Living Cell Shapes. In Linwei Wang, Qi Dou, P. Thomas Fletcher, Stefanie Speidel, Shuo Li. International Conference on Medical Image Computing and Computer Assisted Intervention. Switzerland: Springer Nature Switzerland, 2022. p. 58-67., ISBN 978-3-031-16440-8. doi:10.1007/978-3-031-16440-8_6.BibTeX:@InProceedings{wiesner2022miccai,
title={Implicit Neural Representations for Generative Modeling of Living Cell Shapes},
author={Wiesner, David and Suk, Julian and Dummer, Sven and Svoboda, David and Wolterink, Jelmer M.},
editor={Wang, Linwei and Dou, Qi and Fletcher, P. Thomas and Speidel, Stefanie and Li, Shuo},
booktitle={Medical Image Computing and Computer Assisted Intervention -- MICCAI 2022},
year={2022},
publisher={Springer Nature Switzerland},
address={Cham},
pages={58--67},
isbn={978-3-031-16440-8},
doi={10.1007/978-3-031-16440-8_6}
}
## Acknowledgements
This work was partially funded by the 4TU Precision Medicine programme supported by High Tech for a Sustainable Future, a framework commissioned by the four Universities of Technology of the Netherlands. Jelmer M. Wolterink was supported by the NWO domain Applied and Engineering Sciences VENI grant (18192). We acknowledge the support by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) (Czech-BioImaging Projects LM2023050 and CZ.02.1.01/0.0/0.0/18_046/0016045). This project has received funding from the European High-Performance Computing Joint Undertaking (JU) and from BMBF/DLR under grant agreement No 955811. The JU receives support from the European Union’s Horizon 2020 research and innovation programme and France, the Czech Republic, Germany, Ireland, Sweden and the United Kingdom.
The data set of Platynereis dumerilii embryo cells is courtesy of Mette Handberg-Thorsager and Manan Lalit, who both have kindly shared it with us.
The shape descriptors in the paper were computed and plotted using an online tool for quantitative evaluation, Compyda, available at https://cbia.fi.muni.cz/compyda. We thank its authors Tereza Nečasová and Daniel Múčka for kindly giving us early access to this tool and facilitating the evaluation of the proposed method.
The neural network implementation is based on DeepSDF, MeshSDF, and SIREN.