TRAgen (track generator)
This web page is a supplementary material to the paper TRAgen: A Tool for Generation of Synthetic Time-Lapse Image Sequences of Living Cells presented at 18th International Conference on Image Analysis and Processing (ICIAP 2015) hold in Genova, Italy in September 2015.
Summary
The paper introduces a tool, capable of generating 2D image sequences showing simulated living cell populations together with ground-truth images for evaluation of cell tracking tasks. The simulated events include namely cell motion, cell division, cell death, and cell clustering up to tissue-level density. TRAgen features complete cell cycle with shape changes due to rounding and elongation of a cell just before its division.
It is primarily designed to operate at inter-cellular scope, that is, it predominantly focuses on interaction between cells. It does not handle explicitly any intra-cellular structures such as protein molecules, chromatin territories, or mitochondria. Instead, the implementation provides mean for user to inject his/her own texture to be overlaid over the generated cell mask.
User can control the rate of cell divisions, average speed of cells, and temporal and spatial sampling of the simulation to steer complexity of the tracking task.
It is designed to provide a sequence of 2D images with labelled masks together with lineage trees to provide a complete tracking ground-truth data. We believe that prospective authors of newly developed tracking algorithms may easily add appropriate textures to these masks and tailor appearance of the obtained testing images specifically to the context of their needs, e.g., to phase-contrast or fluorescence microscopy, or to images with different SNR. The generated testing images obtained in this way, may become yet more realistic and thus more relevant for the evaluation.
Sample Sequence
The main aim of this project is to handle movement and inter-cellular interaction of cell digital phantoms. In the following two images (screenshots of selected time-lapse sequences), you can inspect all implemented forces affecting the motion and interaction of individual cells in the population. The left image shows the overall cell population while the right one focuses on four selected cells.
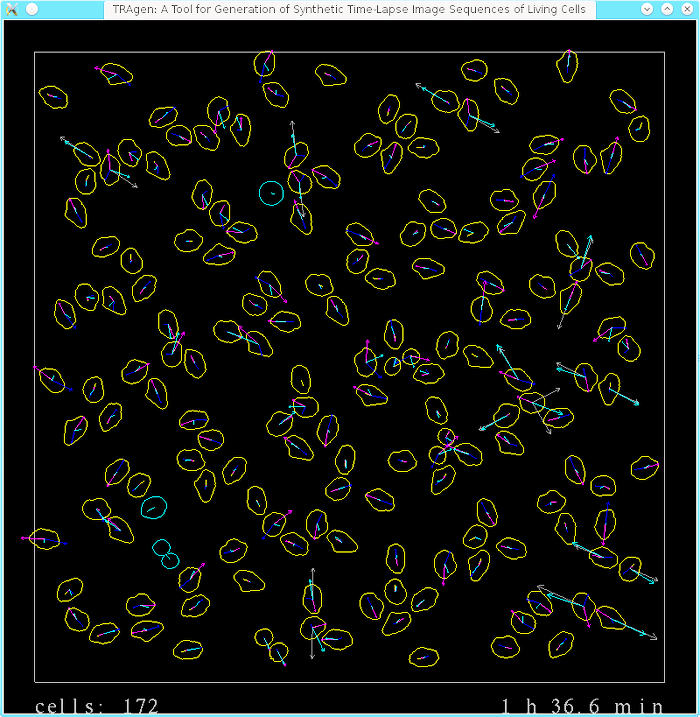
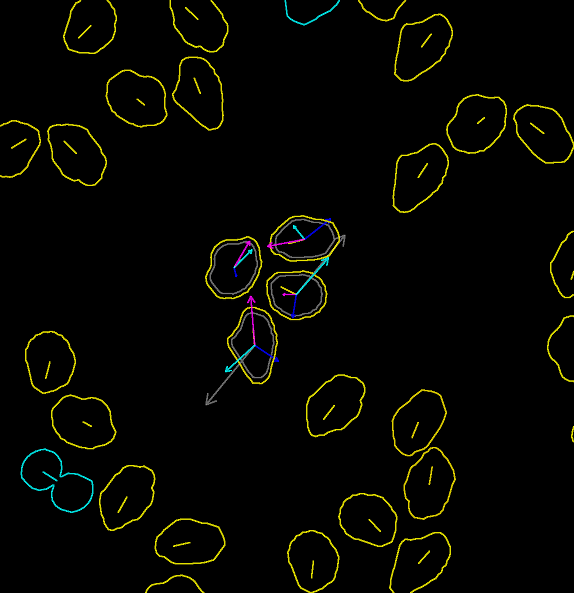
The emphasis on movement and inter-cellular interaction basically boils down to the activity of “pure” displacing and altering shape of cell masks. Thus, the implementation/generation of the content to be filled into the masks as well as outside them, and addition of microscopy and acquisition effects, such as blur, uneven illumination, various kinds of noise, is greatly simplified in the videos below:
-
Videos of cells in phase-contrast microscopy [1 MB] and labelled ground-truth masks [3 MB].
The phase-contrast microscopy video should resemble the Cell Tracking Challenge training dataset “PhC-C2DL-PSC”. To obtain it, the labelled masks were thresholded to values 132 for background and 255 for foreground/cells, value of 109 was inserted into stripes that were eroded by disk shaped SE with 2px radius, and Gauss filtering with sigma of 1px finished the preparation of images. The video with labelled ground-truth images was additionally colour-coded as TRAgen produces originally 16bit grayscale labelled masks.
-
Videos of fluorescence stained cells [138 MB] and labelled ground-truth masks [29 MB].
The fluorescence microscopy video was obtained by mapping pre-processed texture images on the cells (texture images are included in the download archive), smoothed with Gaussian filter, and overlaid with Poisson noise, dark current noise and read-out noise (details can be found in the source code file graphics.cpp). The video with labelled ground-truth images was additionally colour-coded as TRAgen produces originally 16bit grayscale labelled masks.
-
Video showing content of the TRAgen window during simulation [139 MB].
The last video demonstrates some (mainly visualization) features that are available in the TRAgen and that can be used to aid tuning of the parameters of the simulation. Notice the number of cells and time stamp at the bottom, that indicate density of the population (which stabilizes at around 1050 cells at the end of the video) and current displaying rate of the simulation (steps available are 0.1 min, 1 min, 10 min), respectively. TRAgen allows to show (user scaled) various individual forces (or current velocities in magenta) over the whole population or only for mouse-selected cells. Notice the cell outline darkens proportionally to the magnitude of the greatest individual force; yellow cells are in the interphase, blue ones in mitosis. If a single force exceeds both given threshold of magnitude and acting time period, the cell is killed. Later in the video, we intervened the simulation and killed group of cells in the middle to show the ability of the system to grow up to fill the hole. The video in its second half shows proliferating population of cells that reaches tissue-like density.
Download and License
The program is written in C++. It uses OpenGL (with GLSL ver 1.5), GLEW, and GLUT libraries for drawing on the screen and rendering into memory buffers. It uses GNU GSL for random number generations. Last but not least, it uses the CBIA i3dlibs for I/O image formats operations plus some additional image manipulation to obtain (superficially) realistic images of the cells.
Source code with CMake configuration script (developed and test in Linux OS), and input data is available. The application has currently no input parameters and does not allow for modification during run-time. Changing parameters of the simulation is available only by changing the source codes and recompiling them.
The TRAgen (the above provided source code and data) is freely available under the GNU GPL license. Copyright belongs to the authors below and Centre for Biomedical Image Analysis (CBIA) at Masaryk University.
Acknowledgement
This project was funded by Czech Science Foundation, grant No. GA14-22461S.