CytoPacq
CytoPacq is a free web-based service for generating synthetic multi-dimensional images of cells and their populations in optical microscopy with accompanying reference annotations. The aim of the service is to provide an easily-accessible graphical interface for generating benchmark datasets suitable for testing and validation of bioimage analysis methods. Based on the difference from an automatically-generated reference annotation, one can easily compare the quality and accuracy of results of bioimage analysis methods, such as segmentation, tracking, or deconvolution.
Trivia: The web-interface name originates from words “CYTo”, “OPTics”, and “ACQuisition”, where “cyto” originates from Ancient Greek word “kytos”, meaning “cell” in english. The connotation of the name itself is “CYTOmetry PACKage”.
Important Features
- fully 3D output: specimens are simulated in arbitrary resolution with configurable z-step
- time-lapse sequences: ability to generate dynamic sequences with configurable duration of the experiment
- phantom fixation: simulations can be reiterated using previously generated digital phantoms1 to simulate the same scene in different microscopy modes or with different acquisition settings
- real-life resemblance: employed computational models cover the most common phenomena occurring during real acquisition process
- ease of use: easily accessible web-based graphical interface2 with contextual help, emphasizing the most important parameters
- active project: new modules are incorporated as our development on the synthesis of bioimage data advances
1 Each generated digital phantom is unique, due to the random nature of the employed computational models, hence the ability to (re)use it as a base for another simulation, with arbitrary configuration of virtual optical system and virtual acquisition device, is crucial for generating benchmark datasets.
2 Web-interface is working in standard web browsers, i.e. Chrome, Safary, Firefox, Edge, and Opera, with no requirements for additional modules like Java or Flash.
CytoPacq
A web-based simulation framework capable of generating artificial microscopy image data fully in 3D+time. The simulated data imitate the image as if acquired using the fluorescence optical microscope.
Feel free to check this service and use the generated data!
Quick Start
After opening the CytoPacq web-interface in your browser, you are presented with the input form divided into three columns, digital phantom, optical system, and acquisition device, corresponding to the underlying modules. The question mark icon on the left side of every parameter row represents a contextual help, explaining the impact of the parameter and offering useful tips. Contextual help for a particular parameter can be accessed by hovering the mouse cursor over the corresponding icon.
You can begin generating data in just two easy steps (see Fig. 1). First, you have to select the type of digital phantom that you want to simulate, e.g. a single cell with filopodia. And second, you have to click on the “Start simulation” button. Everything else will be handled in the background automatically by the web-interface simulation system.
Note: As the simulation process can take a substantial amount of time, depending on the chosen settings, we do not oblige you to leave the web-interface open in your browser for the whole process. To get back to your simulation later, you can either enter your e-mail (see row marked with an asterisk on Fig. 1) and use the uniquely generated link in the received notification, or you may bookmark the page in your browser.
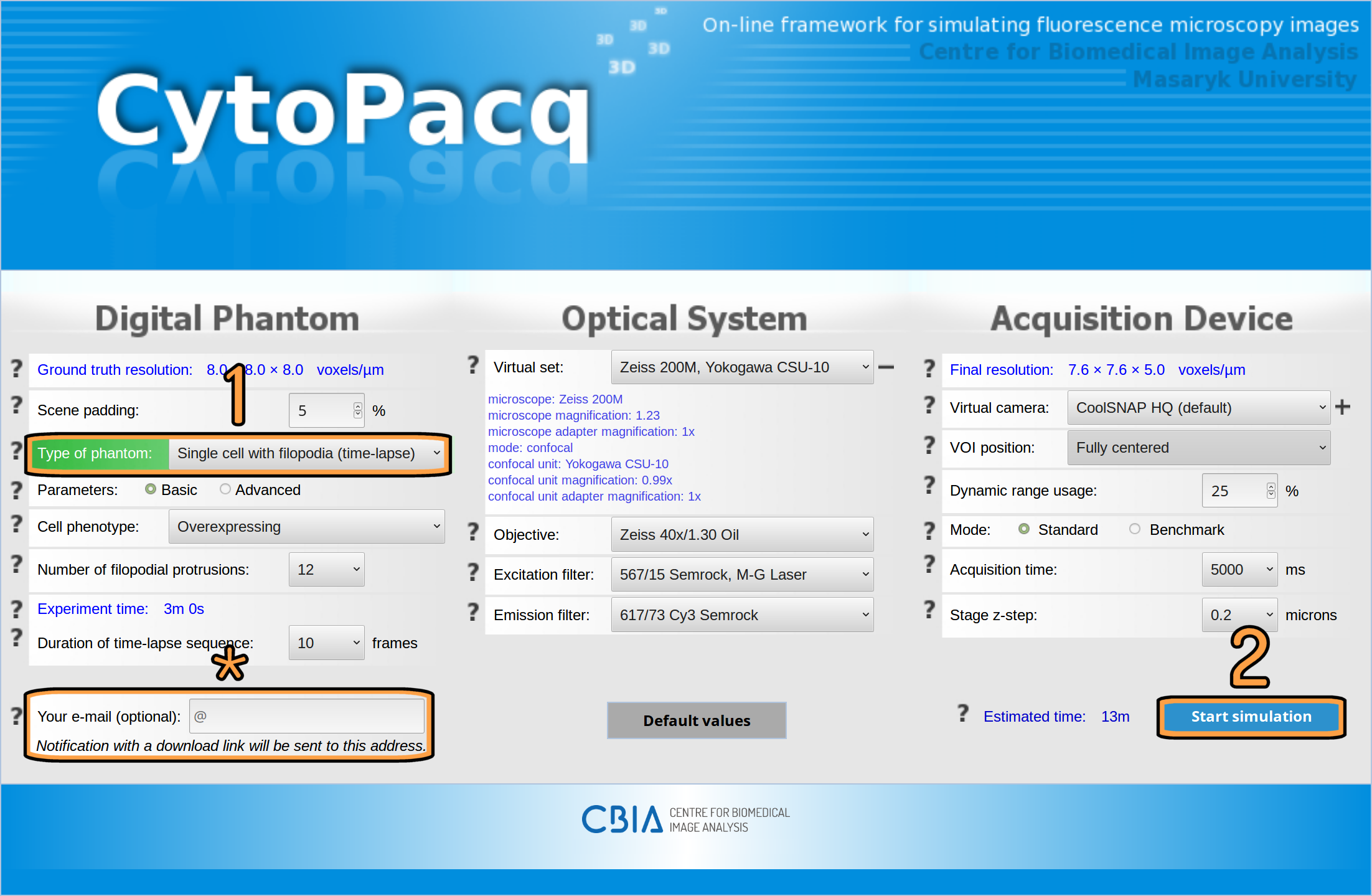
Fig. 1: Configuration form with simulation parameters in the CytoPacq web-interface.
Modules
-
DIGITAL PHANTOM
The following modules handle the generation of 3D digital cell phantoms (spatial objects imitating cells and their components as well as their structure).
- Modeling of static cell populations - CytoGen
Generated ground truth: cell phantom, labeled mask
CytoGen module is capable of generating populations of fixed HL60 cell nuclei, granulocytes, C. elegans, or clusters of human colon tissues.
- Modeling of mitosis - MitoGen
Generated ground truth: cell phantom, labeled mask, cell lineage
MitoGen module implements a model of HL60 cell population that evolves in time. All the cells are modelled fully in 3D+time. They can move and split due to mitosis that is also simulated.
- Modeling of single cells with filopodial protrusions - FiloGen
Generated ground truth: cell phantom, labeled mask
FiloGen module realizes a model of single lung cancer cells with evolving filopodia. The cell is modeled fully in 3D+time.
-
OPTICAL SYSTEM - OptiGen
A module simulating image formation in the optical system, i.e.:
- blurring process occurring in the optical system
- uneven illumination
- virtual excitation/emission filters (in the case of fluorescence imaging)
-
ACQUISITION DEVICE - AcquiGen
A module simulating the phenomena manifesting themselves during image capture using digital image detectors, i.e.:
- dark current signal
- (re)sampling
- fixed pattern noise
- quantification uncertainty (poisson noise)
- amplification (readout noise)
- A/D conversion (quantization)
Workflow
The simulation process (see Fig. 2) begins by specifying desired configuration, which you can easily define by filling the input form in the web-interface (see Fig. 1). This configuration is subsequently passed to the appropriate simulator, depending on the chosen type of digital phantom. The simulation output then consists of synthetic images and accompanying reference annotations.
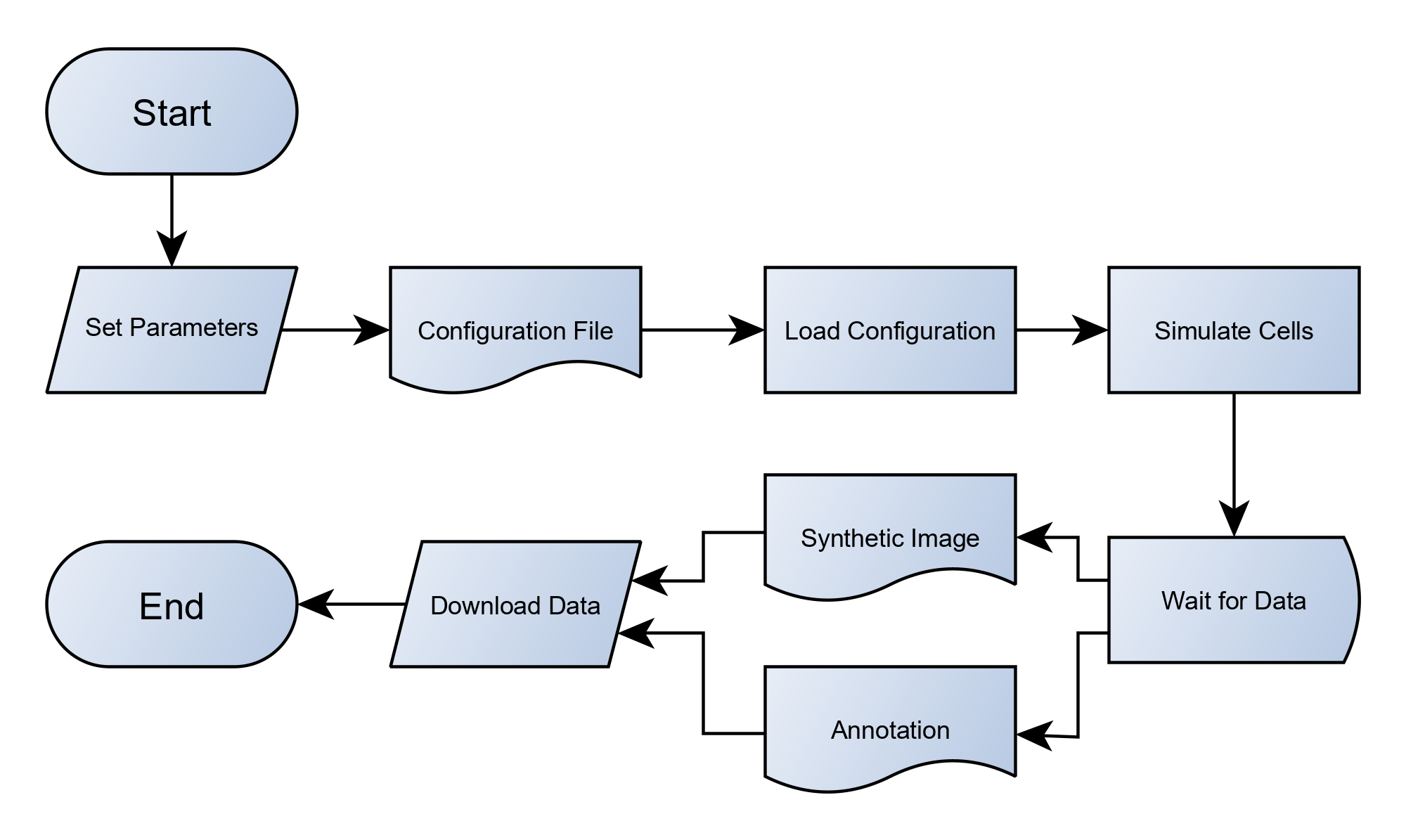
Fig. 2: High-level view (flowchart) of the simulation process.
Obtaining the data
As soon as the simulation is finished, you will be able to obtain the generated data from the download page (see Fig. 3). The download page contains previews of the synthetic images, (selective) downloads of the output data, and links to recommended image viewers.
The image previews correspond to each stage of the simulation process, i.e. digital phantom generation, simulation of optical system, and simulation of acquisition device. You are able to download either complete archive, containing synthetic images of all simulation stages with reference annotations in TIFF or ICS format including generated configuration files, or any of mentioned separately. The recommended viewers, Viewer 3D and Viewer 4D, are our in-house developed tools for convenient browsing of 3D images and their sequences, respectively.
Note: The generated configuration files in INI or JSON format can be used as an input for our simulators, if you wish to install them on your computer and run them offline. More information about particular simulators can be found here.
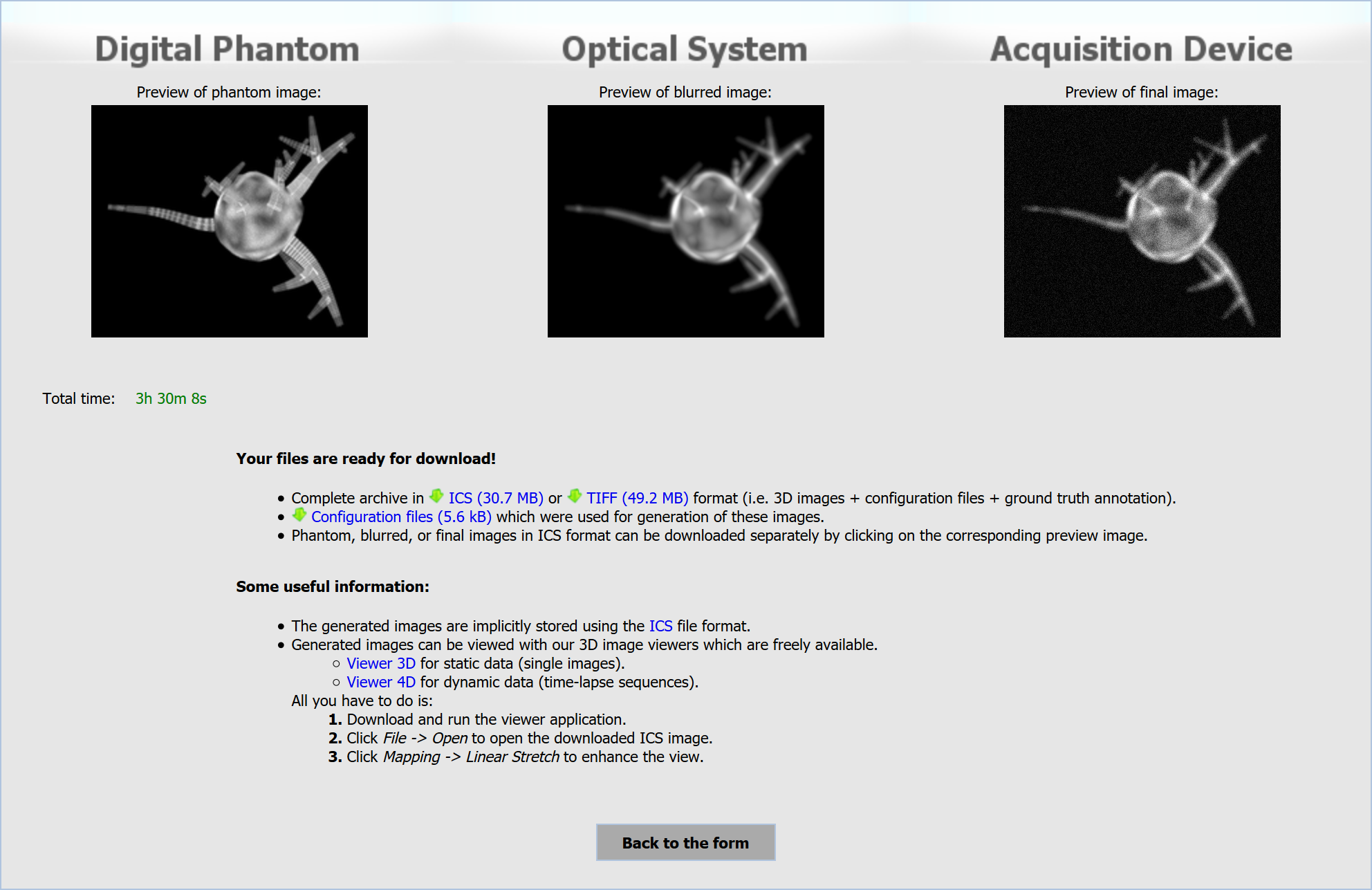
Fig. 3: Download page with a finished simulation.
###Additional Functionality
The following supplementary services are accessible from the main page.
-
Public Benchmark Datasets and their Preconfigured Parameters
Public benchmark datasets, generated by the CytoPacq simulation modules, are available as a part of the Cell Tracking Challenge (CTC), Broad Bioimage Benchmark Collection (BBBC), and Masaryk University Cell Image Collection (MUCIC). Aside from being able to download these public datasets from the main page, user is also offered an option to generate new datasets based on their parameters.
-
My CytoPacq, My Benchmark Datasets
My CytoPacq is a personal area, serving as a hub for services facilitating access and manipulation with user data. My Benchmark Datasets is a part of My CytoPacq, allowing user to access data of their previous simulations at any time directly through the web-interface. Furthermore, the data can be used as a basis for another simulation, i.e. user is able to fix a phantom and generate new data using arbitrary configuration of optical system and acquisition device (see Fig. 4). The phantom fixation is available for microspheres, HL60 nuclei (static and dynamic), granulocyte nucleus, and colon tissue. More services will be added as the development continues. The optional registration is offered to every user who would like to use services in My CytoPacq.
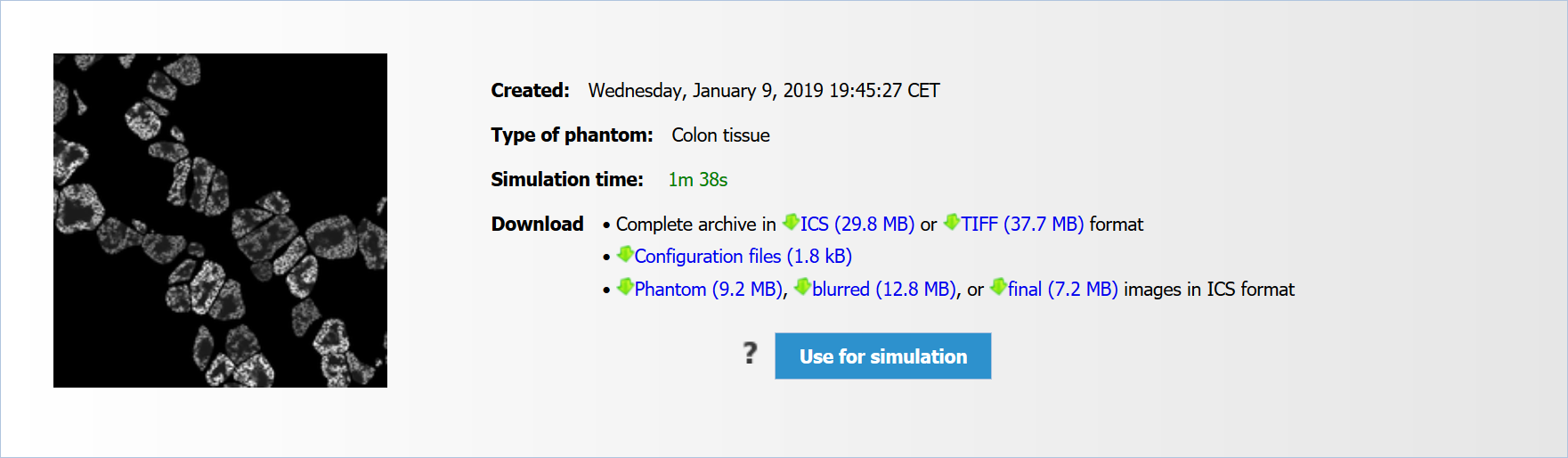
Fig. 4: Access to data of previous simulations in "My CytoPacq".
###Running the Software on Own Hardware
As described in QUICK START section, the simulation has three stages, i.e. simulation of digital phantom, simulation of optical system, and simulation of acquisition device. The CytoPacq web-interface utilizes three simulation modules for digital phantom simulation, CytoGen, OptiGen, and FiloGen. The simulation of optical system and acquisition device is implemented in OptiGen and AcquiGen module, respectively.
All simulation modules are freely available in the form of preconfigured virtual machines, containing the simulation modules and corresponding source codes. More information about running the software on own hardware can be found on the respective web pages of each simulation module (see MODULES section).
Terms of use
The usage of datasets generated by the CytoPacq web-interface (or underlying simulation modules) is endorsed for scientific and educational purposes. The usage for any other purpose (e.g. commercial) is generally possible, but solely with the explicit permission of the authors. All generated data is free of charge and may be used as long as the CytoPacq web-interface is properly acknowledged by using the quotation of this form (may be modified only formally for the needs of individual publication purposes):
The dataset was created using CytoPacq web service[*] (https://cbia.fi.muni.cz/simulator).
[*] Wiesner D, Svoboda D, Maška M, Kozubek M. CytoPacq: A web-interface for simulating multi-dimensional cell imaging. Bioinformatics, Oxford University Press, 2019. ISSN 1367-4803. 2019. doi:10.1093/bioinformatics/btz417.
More information is available here. In case of any questions, please do not hesitate to contact us (see CONTACT section).
Changelog
- VERSION 3.7
- added support for C. elegans phantom and theoretical PSF
- VERSION 3.6
- added web monitoring system
- VERSION 3.5
- included presets of public benchmark datasets
- VERSION 3.4
- simulation archive “My Benchmark Datasets” for registered users
- users are able to use previously generated phantoms from CytoGen and MitoGen modules for another simulation with arbitrary optical system and acquisition device configuration
- VERSION 3.3
- VERSION 3.2
- benchmarking mode for FiloGen module, allowing simulation of multiple configurations of acquisition time and z-step at once
- VERSION 3.1
- FiloGen module updated to generate configuration files in JavaScript Object Notation (JSON)
- VERSION 3.0
- support for dynamic (time-lapse) sequences
- implementation of MitoGen and FiloGen module
- VERSION 2.0
- full integration with MySQL database
- asynchronous communication between front-end and back-end, user is no longer required to leave the page open in browser during the simulation
- VERSION 1.0
- implementation of CytoGen, OptiGen, and AcquiGen module
References
- Wiesner D, Svoboda D, Maška M, Kozubek M. CytoPacq: A web-interface for simulating multi-dimensional cell imaging. Bioinformatics, Oxford University Press, 2019, vol. 35, No 21, p. 4531–4533. ISSN 1367-4803. 2019. doi:10.1093/bioinformatics/btz417.
The following papers describe the methods and principles of data generation used in the modules currently implemented in the CytoPacq web-interface.
Acknowledgement
The progress of work devoted to the simulator was incremental in order to meet actual needs of scientific projects No. 2B06052 and No. LC535 by the Ministry of Education of the Czech Republic, and the scientific projects GBP302/12/G157, GA14-22461S and GJ16-03909 by the Czech Science Foundation.
Credits
- David Wiesner (web development, web admin, web server and database server maintenance)
- David Svoboda (coordinator, image processing, software development)
- Martin Maška (measurement and validation, image processing)
- Vladimír Ulman (optical flow, image processing)
- Dmitry Sorokin (filopodia modeling, image processing)
- Igor Peterlík (physics-based simulations)
- Michal Kozubek (optics, signal sensing)
- Stanislav Stejskal (biology)
- Jaromír Coufal (web design)
- Ondřej Homola (generation of phantom of colon tissues)
- Luděk Matyska jr. (uneven illumination modeling)
- Alena Bednaříková (web monitoring system)
Contact
If you meet some failure or if you like to contact developers, please write an e-mail to cytopacq@fi.muni.cz.




